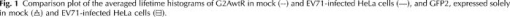|
|
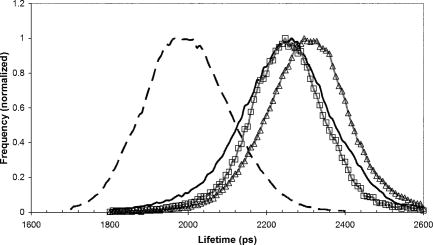
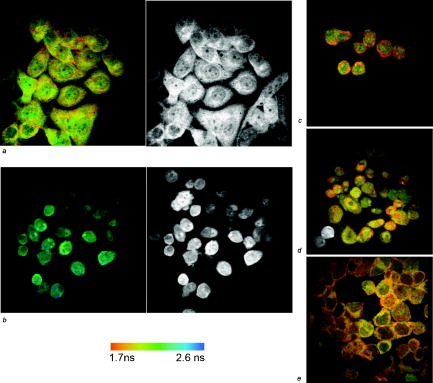
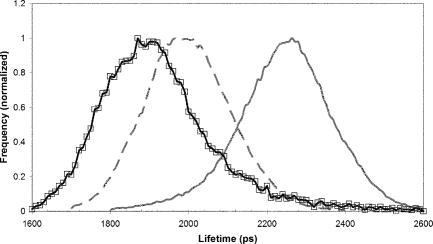
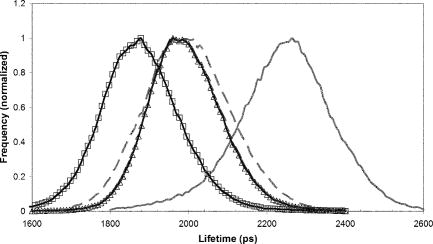
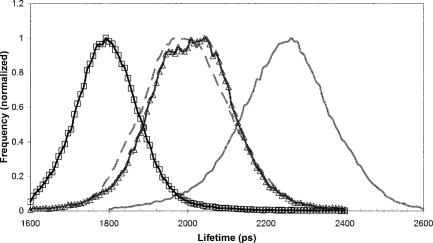
1.IntroductionFirst isolated and characterized1 from cases of neurological disease occurring in California from 1969 to 1973, the enterovirus 71 (EV71) causes seasonal epidemics of hand-foot-and-mouth disease (HFMD) with fatal neurological complications in young children. The virus has been associated with outbreaks in Australia, Brazil, Bulgaria, Hungary, Japan, Malaysia, and the United States. The largest and most severe EV71 epidemic to date occurred2 in Taiwan in 1998. At that time 129,106 cases of HFMD/herpangina were reported, of which 405 cases had severe neurological complications and/or pulmonary edema, and 78 cases of child mortality were claimed. The recent HFMD outbreak in Singapore in October 2000 affected several thousand children and resulted in four deaths.3 EV71, one of 64 enterovirus serotypes, belonging to the Picornaviridae family,4 is a small nonenveloped icosahedral virus with positive single-stranded RNA genome of about 7400 nucleotides5 translated to a single giant polyprotein. Mature proteins are derived in a series of posttranslational cleavages of the precursor protein whereby the initial stages are accomplished by the 2A protease released by the rapid cotranslational cleavage at its own amino terminus.6, 7, 8, 9 In the cytoplasm of a host cell, the performs also a bundle of other activities. Thus, expressed in mammalian cells, caused significant reduction of RNA polymerase type II transcription and, to a lesser extent, DNA transcription.10 Although direct participation of the in these processes has not yet been proven, this evidence demonstrates that like many other viral proteins, is a multifunctional enzyme, which is important for the complete infection cycle to happen. A range of methods are being used for the EV71 infection diagnosis, such as classical serological (including the “golden standard” neutralization test and complement fixation), and the most effective currently reverse transcription polymerase chain reaction (RT-PCR), Southern blot, and others, based on the detection of viral nucleic acids. Approaches based on microscopy techniques have demonstrated high efficiency and advantages in diagnosis speeds as compared to the classical methods.11, 12 Recent advancements in photonics technologies further increased such opportunities. Examples include applications of single-molecule imaging13 and fluorescence correlation spectroscopy14, 15 (FCS) for virus detection. Here we propose another novel approach for the detection of EV71 infection, based on the fluorescence resonance energy transfer (FRET) resolved by fluorescence lifetime imaging microscopy (FLIM). FRET is a quantum mechanical process between two fluorophores whereby energy from an excited donor molecule is transferred to the acceptor molecule by means of a near-field dipole-dipole coupling.16 The efficiency of the process varies with the sixth power of the intermolecular separation and is usually described by means of Forster radius (a distance at which coupling efficiency reaches 50%), typically within the range of . The depletion of the excited state population of the donor at FRET reduces both its fluorescence intensity and lifetime,17 which is an average time the molecule stays in an excited state before returning to the ground state. A method to map the lifetimes distribution on microscopic images, recently FLIM has become a well-established technique to investigate energy transfer processes in cells.18, 19 As compared to the fluorescence intensity-based measurements, FLIM does not depend on the concentration of the fluorophores and is less affected by side effects, such as background noise, spectral hole burning, and photobleaching. Among all lifetime imaging techniques, the time-correlated single-photon counting (TCSPC) has demonstrated the highest recording efficiency and time resolution.20, 21 Mature and reliable, TCSPC is based on the repetitive excitation of a fluorophore, recording of the single photons emitted, measurement of the detection times and reconstruction of the wave form from the individual time measurements.22, 23 Despite alternative and possibly faster recording method reported earlier,24, 25 less accuracy is generally achieved when compared with that of the TCSPC. For the detection of EV71 by means of FLIM/FRET, a genetically modified cell line was created by the transfection of HeLa cells with a plasmid, encoding the FRET pair of donor green fluorescent protein F64L mutant (GFP2) and acceptor DsRed2, linked by a 12-amino-acid-long spacer, whereby the sequence of the link was a cleavage recognition site for the enteroviral . Expressed in cells, the fusion protein, referred to as G2AwtR, served as a reporter molecule for the maturation of enteroviral polyprotein. Upon the initial proteolytic processing and release of the to the cytoplasm, the protease clove the linker between the fluorophores, thus causing FRET disruption and hence increase of the GFP2 lifetime, detected by the TCSPC technique. 2.Materials and Methods2.1.Plasmid ConstructsA recombinant plasmid pG2AwtR composed of the GFP2 and DsRed2 genes with a linker that encodes the predicted EV71 cleavage motif (RTAITTL↓RKFGQ, where the arrow indicates the scissile bond) was constructed. For the mutant control the pG2AwtR has been modified with the substitution in the position of the cleavage motif. pG2AwtR and pG2AmutR plasmids were separately transfected into HeLa cells and subjected to Zeocin selection, followed by a limiting dilution to obtain stable clones, referred to as HeLa-G2AwtR and HeLa-G2AmutR, respectively. 2.2.Cells Infection, Fixation, and PreparationHeLa-G2AwtR and HeLa-G2AmutR cells were subject to infection by EV71 (BrCr strain) or herpes simplex virus (HSV, KOS strain) at a multiplicity of infection for , or left mock-infected. Cells were washed with phosphate-buffered saline (PBS), fixed with formaldehyde (3%) for and sealed in a mounting medium (1:1 ratio of glycerol plus PBS) prior to the lifetime measurements. For apoptosis induction cells were treated with cisplatin at 70% confluence prior to fixation. 2.3.Fluorescence Lifetime ImagingTime-domain FLIM was performed with a 1.45 numerical aperture (NA) PlanApochromat oil objective lens (Olympus Corp.) on a modified two-photon laser scanning microscope (FV300 with the IX71 inverted microscope, Olympus Corp.). Samples were excited at (two photons) by a mode-locked femtosecond Ti:sapphire Mira F-900 laser, pumped by a solid state cw Verdi laser (both from Coherent Inc.) and operated at a frequency. The laser beam was fed into the scanning unit with high-precision alignment. The scanning speed of the FV300 was controlled externally by a function generator (AFG310, Tektronix Inc.) so as to be synchronized with the TSCPC-based multichannel scalar to record the FLIM images. Fluorescence photons were detected in a nondescanned mode by a photon-counting photomultiplier (H7422P-40, Hamamatsu Photonics K.K.). Time-resolved detection was conducted by the SPC-830 PC board (Becker&Hickl GmbH) with the repetition rate of the Ti:sapphire laser serving as the time base. A green bandpass filter of (Edmund Optics Inc.) was used in the detection channel for the GFP lifetime measurements along with a shortpass IR cut-off filter. All the images were taken at resolution with the acquisition times in the range of to reach sufficient photon counts for statistically reliable fitting. 2.4.FLIM Data AnalysisData analysis via model function fitting along with the instrument response function (IRF) deconvolution and color coding was conducted with the commercially available SPCImage v2.8 software package (Becker&Hickl GmbH). Fluorescence decay in each pixel of the analyzed lifetime images may result from a few fluorophore molecules and vary with their different states and conformations. To describe the composite decays the software utilizes a multiexponential function with offset correction for the ambient light and/or dark noise. On the prior convolution with the IRF, the model function is fitted to the actual data by iterative reconvolution: where is the instrument response, which in our analysis was automatically calculated by software (approximation by a Gaussian function and the width adjustment for the best fit to the rising edge of the decay functions), is the baseline offset, and represents the main components of the resulting fluorescence decay. The intensity coefficients have relative amplitudes and demonstrate the correlation between components fractions populations. Due to the extremely low duration as compared to the temporal resolution of the photomultiplier used, the scattering parameter was not considered for the purpose of this experiment and was fixed with the meaning of 0.The reduced goodness-of-fit parameter was calculated by where is the actual experimental data, calculates decay as already described, is the number of the data (time) points, and is the number of the model parameters.26 The was minimized by the Levenberg-Marquardt search algorithm.The main comparison analysis of G2AwtR lifetime histogram, recorded in the GFP channel, with that of mutant virus specificity and apoptosis controls was conducted by fitting the experimental data to a single-exponential decay model. The autofluorescence from HeLa cells and bleedthrough from dsRed2 were found negligible and were reduced by increasing the threshold parameter at the analysis of lifetime images. The deviation in measured lifetime was obtained statistically and does not represent the width of IRF. 3.Results3.1.Indicating the EV71 Virus InfectionOne of the main indicators of FRET occurring is a shortening of the donor lifetime in the presence of an acceptor. To obtain a reference point for further measurements, the lifetime of the GFP2 was first characterized under sole expression in HeLa cells. Recorded from the mock cells, GFP2 lifetime was equal to , whereas at virus infection the lifetime was shifted slightly to the shorter range and resulted in an averaged histogram with the peak at [Figs. 1 and 2a ]. Fig. 2Fluorescence intensity decays recorded from (a) GFP2, solely expressed in HeLa cells; (b) mock HeLa-G2AwtR; and (c) HeLa-G2AwtR, infected with EV71. 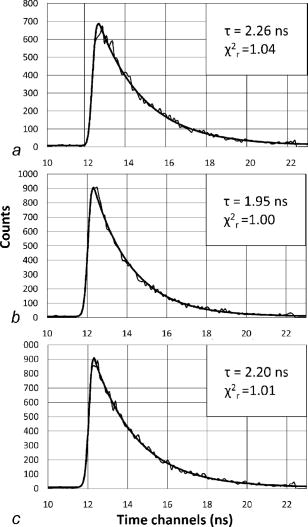 Results obtained at the measurements from HeLa-G2AwtR are presented in Figs. 1, 2b, 2c, 3b, 3c . Lifetime recording in the GFP-channel, taken from the mock-infected cells, resulted in a histogram with a plateau-shaped top, centered at . For the cells infected with the EV71 virus, we observed a shift to longer lifetimes, reaching the peak of . As demonstrated on the comparison plot, on infection the histogram obtained from the G2AwtR lifetime measurements follows the pattern of that recorded from the GFP2 expressed solely, indicative of the fluorophores separation and thus FRET signal disruption as a result of the linker cleavage by the viral . 3.2.Effect of Cytopathic Events on the Reporter Molecule IntegrityBesides the changes in the GFP2 lifetime measured from HeLa-G2AwtR, virus activity also caused a characteristic cytopathic effect (CPE), altering the morphology of cells as well, as observed in our experiments [Fig. 3b]. Earlier it was reported that one of the CPE effects, caused by EV71 is the launch of a programmed cell death, or apoptosis, whereas the protease activity was demonstrated as possible primary initiator of the cascade of reactions, leading to the cell self-termination machinery initiation.27, 28 The apoptotic pathway involves a family of aspartate-specific cysteinyl proteases (cysteine aspartases or caspases), activated by proteolytic cleavage. This activation leads to the processing of various cytoplasmic and nuclear targets by a subclass of downstream effectors termed “executioners.”29, 30 To investigate involvement of apoptotic proteolytic machinery to the disruption of G2AwtR protein, HeLa cells were treated with cis-diamminedichloroplatinum(II) (cisplatin)—a known biochemical inducer of apoptosis. The results, presented in Figs. 3c and 4 , indicate no FRET disruption on apoptosis induction. On the contrary, the GFP lifetime in cisplatin-treated samples tends to shift to shorter values . 3.3.Specificity of the Method to EV71 DetectionSpecificity of the FRET disruption and direct dependence of the linker cleavage on release is demonstrated with two additional series of control measurements. In one case, the linker between the fluorophores was substituted with a mutant variant G2AmutR. The mutation, carried out as described in Sec. 2 affected the cleavage site in a way to prevent its recognition by the . Although the change of HeLa-G2AmutR morphology at infection was similar to that of HeLa-G2AwtR, no lifetime decrease was recorded, indicating no FRET disruption [Figs. 3d and 5 ]. As in the case with the cisplatin-induced apoptosis, analysis of fluorescent decays from these samples revealed a decrease of lifetime as compared to the mock cells by . To check the authenticity of the G2AwtR to the detection of polioviruses on the example of EV71, we tested an alternative virus infection effect on our construct. For this purpose we infected our HeLa-G2AwtR cells with the HSV. A member of a Herpesviridae family, HSV differs from EV71 in the proteins encoded and main infection pathways. The GFP2 in the reporter G2AwtR exhibited reduction of lifetime at HSV infection, similar to that observed for the lifetime, recorded from apoptotic HeLa-G2AwtR and EV71-infected HeLa-G2AwtR, showing integrity of the reporter in cells [Figs. 3e and 6 ]. 3.4.Double-Exponential AnalysisDouble-exponential analysis was applied to reveal the distribution of interacting and noninteracting populations in the samples and map the actual separation of the lifetimes. We found that the fraction of noninteracting population is negligible, showing that the majority of the expressed reporter molecules have been recognized by the and cleft (data not shown). Lifetime color mapping also revealed that not all the cells underwent the FRET disruption at the same extent, which might be a result of nonsimultaneous infection. 4.Discussion4.1.G2AwtR Lifetime BehaviorExpressed in cells, the G2AwtR construct spread evenly over the cells both in cytoplasm and in nucleus, whereas in nucleus the lifetime appeared to be slightly longer than in cytoplasm (Fig. 7 ), which we assume might result from a pH difference. Besides the intrinsic characteristics, the fluorescence decay of fluorophores depends in a measurable way on the environment, such as viscosity, refractive level, interactions with other molecules, and pH.31 Ambient pH has been shown to influence spectral characteristics of wild-type GFP and a number of its mutants, suggesting pH sensitivity for the entire class of these proteins.32, 33, 34 Some authors35, 36, 37 have previously shown that nuclear pH was above that of the cytosol, which is explained by the permeability barrier to ions by nuclear membrane. This might have been the main cause of the wider standard deviation for the lifetime measured from mock-infected cells (see Table 1 ) and thus broader, plateau-shaped histograms. Fig. 7Lifetime histograms and color-coded images of the G2AwtR construct expressed in cytoplasm and nucleus: (a) the color-coded image of the specimen shows a clear contrast between the nucleus (blue) and cytoplasm (orange, green); (b) selection area for the G2AwtR lifetime histogram recording from the cytoplasm (—); and (c) selection area for the G2AwtR lifetime histogram recording from nucleus (--). (Color online only.) 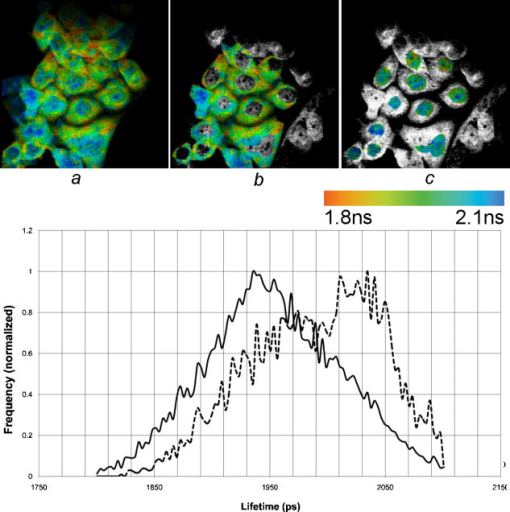 Table 1Averaged lifetime histograms peaks and standard deviation values.
Among other events at apoptosis, which has been shown to be initiated at EV71 infection, different research groups have reported perturbation of the nuclear envelope and clustering of nuclear pores, which leads to the destruction of the permeability barrier.38, 39, 40 This is in good compliance with our results, obtained from the infected cells—we found that on EV71 infection and change of the cell morphology, characteristic for apoptosis, the difference in lifetime between cytoplasm and nucleus is reduced or completely equalized. The concept is supported with the less deviation of the data in infected cells and sharper peak in averaged histograms obtained. Contributions to this suggestion are the fact that HSV induces apoptosis in cells and a previous report stating that intracellular acidification is one of the apoptosis features, whereas the cytoplasmic pH level, normally exhibiting 7.4, decreases41 to the level of 6.6 to 6.7.46 Taking these data into consideration, we suggest recognizing intracellular acidification as a main reason for the decrease in lifetime recorded in the green channel from mutant FRET complex infected with EV71 and wild-type FRET complex in cells infected by HSV. We admit, however, that this issue was out of the scope of the given research and we did not conduct the infection by HSV (reported time to produce apoptotic morphological changes) to check this suggestion. 4.2.Method Advantages and Application ScopeSeveral protein constructs that put FRET-interacting fluorophores on either side of a protease cleavage recognition site were reported earlier for different applications of FLIM/FRET microscopy, including, but not limited to, the study of proteases activity,42, 43 such as caspases activities during apoptosis,44, 45 and FRET efficiency measurements.46 Having developed a construct with the cleavage recognition site for EV71-specific and applying it with FLIM/FRET microscopy, we demonstrated an alternative imaging-based approach with high sensitivity and efficiency for the virus infection detection. The series of experiments conducted demonstrated that the G2AwtR construct reports specifically the appearance of the in the cytoplasm and is not affected by the internal protein machinery even during the critical apoptotic reactions and a range of other CPEs caused by virus infection. Based on the sensitivity to a viral protein, the approach proposed does not depend on the detection of antigens and/or antibodies and is advantageous as compared to the immunofluorescent methods of serological detection, not applicable with untypable viruses and antigenic drifts. As compared to the direct electron microscopy methods, the approach described provides higher efficiency and speed of detection; no special operations are required for the sample preparation. The FLIM/FRET microscopy is independent of the fluorophore concentration, so that even small changes in signal can be detected and thus just a small number of viruses is sufficient for the infection detection. The proposed method is not applicable to all viral infection detection, provided by serological and viral nucleic acid detection methods, and does not enable us to identify different serotypes of the same virus, which may be important to recognize serotype-specific clinical illness. However, the protease-specific lifetime-based detection method still applies to a range of viruses, encoding a specific protease, whereas definition of the serotypes does not change the clinical management of patients. Thus, the development of a cell line with the reporter construct described, its infection with the isolated viruses, and further observation with FLIM/FRET microscopy can be applied as a fast, sensitive, and efficient method for the detection of certain viral infection. AcknowledgmentsThis work was financially supported by the Ministry of Education, Taiwan (Aim for the Top University Plan, Grant No. 95A-CT8G01), and National Science Council, Taiwan (under Grants NSC95-2112-M010-001 and NSC95-3112-B-010-015). ReferencesN. J. Schmidt,
E. H. Lennette, and
H. H. Ho,
“An apparently new enterovirus isolated from patients with disease of the central nervous system,”
J. Infect. Dis., 129 304
–309
(1974). 0022-1899 Google Scholar
M. Ho,
E. R. Chen,
K. H. Hsu,
S. J. Twu,
K. T. Chen,
S. F. Tsai,
J. R. Wang, and
S. R. Shih,
“An epidemic of enterovirus 71 infection in Taiwan,”
N. Engl. J. Med., 341 929
–935
(1999). 0028-4793 Google Scholar
K. Ahmad,
“Hand, foot and mouth disease outbreak reported in Singapore,”
Lancet, 356 1338
(2000). 0140-6736 Google Scholar
A. M. Q. King,
F. Brown,
P. Christian,
T. Hovi,
T. Hyypiä,
N. J. Knowles,
S. M. Lemon,
P. D. Minor,
A. C. Palmenberg,
T. Skern, and
G. Stanway,
“Family Picornaviridae,”
Virus Taxonomy. Seventh Report of the International Committee for the Taxonomy of Viruses, 657
–673 Academic Press, San Diego
(2000). Google Scholar
B. A. Brown and
M. A. Pallansch,
“Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus,”
Virus Res., 39 195
–205
(1995). 0168-1702 Google Scholar
M. Ryan and
M. Flint,
“Virus-encoded proteinases of the picornavirus super-group,”
J. Genet. Virol., 78 699
–723
(1997). Google Scholar
M. J. H. Nicklin,
H. Toyoda,
M. G. Murray, and
E. Wimmer,
“Proteolytic processing in the replication of polio and related viruses,”
Nat. Biotechnol., 4 33
–42
(1986). 1087-0156 Google Scholar
H. Toyoda,
M. J. H. Nicklin,
M. G. Murray, and
E. Wimmer,
“Proteolytic processing of the poliovirus polyprotein by two virus-encoded proteinases,”
Protein Engineering: Applications in Science Medicine and Industry, 319
–337 Academic Press, New York
(1986). Google Scholar
A. C. Palmenberg,
“Proteolytic processing of picornaviral polyprotein,”
Annu. Rev. Microbiol., 44 603
–623
(1990). 0066-4227 Google Scholar
M. V. Davies,
J. Pelletier,
K. Meerovitch,
N. Sonenberg, and
R. Kaufman,
“The effect of poliovirus proteinase 2Apro expression on cellular metabolism,”
J. Biol. Chem., 266
(22), 14714
–14720
(1991). 0021-9258 Google Scholar
W.-T. Liu,
J.-R. Sun,
C.-H. Lin,
R.-L. Kuo, and
S.-H. Kung,
“An indicator cell assay for detection of human cytomegalovirus based on enhanced green fluorescent protein,”
J. Virol. Methods, 96
(1), 85
–92
(2001). 0166-0934 Google Scholar
S.-H. Kung,
Y.-C. Wang,
C.-H. Lin,
“Rapid diagnosis and quantification of herpes simplex virus with a green fluorescent protein reporter system,”
J. Virol. Methods, 90 205
–212
(2000). 0166-0934 Google Scholar
G. Seisenberger,
M. U. Ried,
T. Endreß,
H. Büning,
M. Hallek, and
Ch. Bräuchle,
“Real-time single-molecule imaging of the infection pathway of an adeno-asssociated virus,”
Science, 294
(5548), 1929
–1933
(2001). 0036-8075 Google Scholar
N. Walter,
P. Schwille, and
M. Eigen,
“Fluorescence correlation analysis of probe diffusion simplifies quantitative pathogen detection by PCR,”
Proc. Natl. Acad. Sci. U.S.A., 93 12805
–12810
(1996). https://doi.org/10.1073/pnas.93.23.12805 0027-8424 Google Scholar
F. Oehlenschläger,
P. Schwille, and
M. Eigen,
“Detection of HIV-1 RNA by nucleic acid sequence-based amplification combined with fluorescence correlation spectroscopy,”
Proc. Natl. Acad. Sci. U.S.A., 93 12811
–12816
(1996). 0027-8424 Google Scholar
T. Förster,
“Intermolecular energy migration and fluorescence,”
Ann. Phys. (N.Y.), 6 55
–75
(1948). 0003-4916 Google Scholar
W. Becker,
K. Benndorf,
A. Bergmann,
C. Biskup,
K. Konig,
U. Tirplapur, and
T. Zimmer,
“FRET measurement by TCSPC laser scanning microscopy,”
Proc. SPIE, 4431 94
–98
(2001). 0277-786X Google Scholar
S. M. Ameer-Beg,
M. Peter,
M. D. Keppler,
S. Prag,
P. R. Barber,
T. C. Ng, and
B. Vojnovic,
“Dynamic imaging of protein-protein interactions by MP-FLIM,”
Proc. SPIE, 5700 152
–161
(2005). 0277-786X Google Scholar
H. Wallrabe and
A. Periasamy,
“Imaging protein molecules using FRET and FLIM microscopy,”
Curr. Opin. Biotechnol., 16 19
–27
(2005). 0958-1669 Google Scholar
K. Carlsson and
J. P. Philip,
“Theoretical investigation of the signal-to-noise ratio in fluorescence lifetime imaging techniques,”
Proc. SPIE, 4622 70
–78
(2002). 0277-786X Google Scholar
J. P. Philip and
K. Carlsson,
“Theoretical investigation of the signal-to-noise ratio in fluorescence lifetime imaging,”
J. Opt. Soc. Am. A, 20 368
–379
(2003). 0740-3232 Google Scholar
W. Becker, Advanced Time-Correlated Single-Photon Counting Techniques, Springer, Berlin, Heidelberg, New York
(2005). Google Scholar
D. V. O’Connor and
D. Phillips, Time-Correlated Single Photon Counting, Academic Press, London
(1984). Google Scholar
E. P. Buurman,
R. Sanders,
A. Draaijer,
H. Gerritsen,
J. J. F. van Ween,
P. M. Houpt, and
Y. K. Levine,
“Fluorescence lifetime imaging using a confocal laser scanning microscope,”
Scanning, 14 155
–159
(1992). 0161-0457 Google Scholar
H. C. Gerritsen,
N. A. H. Asselbergs,
A. V. Argonskaia, G. J. H. M. van Sark,
“Fluorescence lifetime imaging in scanning microscopes: acquisition speed, photon economy and lifetime resolution,”
J. Microsc., 206 218
–224
(2002). https://doi.org/10.1046/j.1365-2818.2002.01031.x 0022-2720 Google Scholar
J. Lakowicz,
“Time-domain lifetime measurements,”
Principles of Fluorescence Spectroscopy, 118
–121 Kluwer Academic/Plenum Publishers, New York
(1999). Google Scholar
R.-L. Kuo,
S.-H. Kung,
Y.-Y. Hsu, and
W. T. Liu,
“Infection with enterovirus 71 or expression of its 2A protease induces apoptotic cell death,”
J. Genete. Virol., 83 1367
–1376
(2002). Google Scholar
D. Goldstaub,
A. Gradi,
Z. Bercovitch,
Z. Grosmann,
D. Y. Nophar,
S. Luria,
N. Sonenberg, and
Ch. Kahana,
“Poliovirus 2A potease induces apoptotic cell death,”
Mol. Cell. Biol., 20
(4), 1271
–1277
(2000). 0270-7306 Google Scholar
D. R. Green,
“Apoptotic pathways: the roads to ruin,”
Cell, 94 695
–698
(1998). 0092-8674 Google Scholar
G. S. Salvesen and
V. M. Dixit,
“Caspases: intracellular signaling by proteolysis,”
Cell, 91 443
–446
(1997). 0092-8674 Google Scholar
B. Valeur, Molecular Fluorescence: Principles and Applications, Wiley-VCH, Weinheim
(2001). Google Scholar
W. W. Ward,
H. J. Prentice,
A. F. Roth,
C. W. Cody, and
S. C. Reeves,
“Spectral perturbations of the Aequorea green-fluorescent protein,”
Photochem. Photobiol., 35 803
–808
(1982). 0031-8655 Google Scholar
G. H. Patterson,
S. M. Knobel,
W. D. Sharif,
S. R. Kain, and
D. W. Piston,
“Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy,”
Biophys. J., 73 2782
–2790
(1997). 0006-3495 Google Scholar
M. Kneen,
J. Farinas,
Y. Li, and
A. S. Verkman,
“Green fluorescent protein as a noninvasive intracellular pH indicator,”
Biophys. J., 74
(3), 1591
–1599
(1998). 0006-3495 Google Scholar
O. Seksek and
J. Bolard,
“Nuclear pH gradient in mammalian cells revealed by laser microspectrofluorimetry,”
J. Cell. Sci., 109
(1), 257
–262
(1996). 0021-9533 Google Scholar
A. Masuda,
A. Oyamada,
T. Nagaokaa,
N. Tateishic, and
T. Takamatsu,
“Regulation of cytosol-nucleus pH gradients by exchange mechanism in the nuclear envelope of neonatal rat astrocytes,”
Brain Res., 807 70
–77
(1998). 0006-8993 Google Scholar
P. N. Dubbin,
S. H. Cody, and
D. A. Williams,
“Intracellular pH mapping with SNARF-1 and confocal microscopy: pH gradients within single cultured cells,”
Micron, 24 581
–586
(1993). 0968-4328 Google Scholar
J. D. Robertson,
S. Orrenius, and
B. Zhivotovsky,
“Review: nuclear events in apoptosis,”
J. Struct. Biol., 129
(2–3), 346
–358
(2000). 1047-8477 Google Scholar
B. Buendia,
A. Santa-Maria, and
J. C. Courvalin,
“Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis,”
J. Cell. Sci., 112 1743
–1753
(1999). 0021-9533 Google Scholar
M. Kihlmark,
G. Imreh, and
E. Hallberg,
“Sequential degradation of proteins from the nuclear envelope during apoptosis,”
J. Cell. Sci., 114 3643
–3653
(2001). 0021-9533 Google Scholar
R. A. Gottlieb,
“Cell acidification in apoptosis,”
Apoptosis, 1 40
–48
(1995). Google Scholar
R. Heim and
R. Y. Tsien,
“Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer,”
Curr. Biol., 6
(2), 178
–182
(1996). https://doi.org/10.1016/S0960-9822(02)00450-5 0960-9822 Google Scholar
R. D. Mitra,
C. M. Silva, and
C. C. Youvan,
“Fluorescence resonance energy transfer between blue-emitting and red-shifted excitation derivatives of the green fluorescent protein,”
Gene, 173 13
–17
(1996). 0378-1119 Google Scholar
N. P. Mahajan,
D. C. Harrison-Shostak,
J. Michaux, and
B. Herman,
“Novel mutant green fluorescent protein protease substrates reveal the activation of specific caspases during apoptosis,”
Chem. Biol., 6 401
–409
(1999). https://doi.org/10.1016/S1074-5521(99)80051-9 1074-5521 Google Scholar
K. Q. Luo,
V. C. Yu,
Y. Pu, and
D. C. Chang,
“Application of the fluorescence resonance energy transfer method for studying the dynamics of caspase-3 activation during UV-induced apoptosis in living HeLa cells,”
Biochem. Biophys. Res. Commun., 283 1054
–1060
(2001). https://doi.org/10.1006/bbrc.2001.4896 0006-291X Google Scholar
S. Ganesan,
S. M. Ameer-beg,
T. T. C. Ng,
B. Vojnovic, and
F. S. Wouters,
“A dark yellow fluorescent protein (YFP)-based resonance energy-accepting chromoprotein (REACh) for Förster resonance energy transfer with GFP,”
Proc. Natl. Acad. Sci. U.S.A., 103
(11), 4089
–4094
(2006). 0027-8424 Google Scholar
|