|
|
1.IntroductionOptical imaging techniques such as multiphoton and confocal microscopy have greatly contributed to our understanding of biological processes in whole organisms. Recent advances in imaging techniques and hardware components have enabled higher resolution imaging at the subcellular level, greater coverage, and superior signal to noise ratios. While such achievements have resulted in unique opportunities to explore biology in living systems, high resolution in vivo imaging continues to be challenged by problems such as insufficient tissue penetration, difficulties in physically accessing organs within a living animal, and tissue movement. Penetration depth and internal access could in part be addressed by using multiphoton microscopy or miniature objectives.1 Tissue movement, however, is unavoidable as it is the result of vital physiological processes such as respiration and cardiovascular activity. Movement arising from respiration is particularly extensive and has effects on each and every organ of the body and every organ of the body such as tibila anterior,2 lung,3 kidney,4 spinal cord,5 and brain.6,7 Because the time taken to acquire an image is typically the same as or longer than a breathing cycle (approx. 400 ms), respiration can have a very detrimental effect on image acquisition, causing severe image distortions and unstable imaging conditions.2 Faster image acquisition would be one way to overcome this problem, but would likely come at the expense of image quality (high signal-to-noise ratios and/or reduced image sizes). Recently, considerable research has been directed towards developing strategies to overcome motion artifacts.2–11 Unlike previous attempts, the method presented here does not require the construction of an expensive mechanical system4,5 nor does it sacrifice image quality in order to achieve faster image acquisition.8,9 Rather, the only equipment required is a custom made mechanical holder and a commercial ventilator; this setup can be easily applied to all commercial laser scanning microscopes (LSM) without any specific hardware modification. The technique described is based on a dual strategy. First, the tissue to be imaged is softly compressed using the custom-made mechanical holder; this acts to significantly reduce tissue motion in a similar manner as used for beating heart-surgery12 while at the same time it introduces reproducibility in the tissue displacement. The miniaturized mechanical holder is also designed to allow access to organs deep within the mouse and to fine tune its position relative to the imaged organs. Furthermore, the design of the holder is such that it prevents motion along in-plane directions. Second, we implemented a retrospective breathing-gated acquisition modality, performing selective image extraction triggered to a particular phase of the respiratory cycle. The presence of the stabilizer, which introduces position reproducibility of the sample over time, guarantees that all gated images will present a high degree of correlation. By adjusting both the microscope scanning parameters and the breathing frequency, through the use of a mechanical ventilator, we then extract and combine all parts within a sequence of images which belong to the same phase of the respiratory cycle. Final reconstructions are artifact free and representative of actual optical sectioning planes within the tissue. Here, we demonstrate the advantages of this imaging methodology by imaging a designed fluorescent phantom, as well as a mouse kidney in vivo. 2.Materials and Methods2.1.Animal ProtocolsUbiquitin/green fluorescent protein (GFP) expressing mice (C57BL/6-Tg(UBC-GFP)30Scha/J strain, Jackson Laboratories) were anesthetized by mask ventilation of isoflurane (vaporized at 1–2% concentration). Body temperature was maintained at 37.5 °C and mice were placed in a lateral position. To stabilize respiration, animals were intubated and ventilated with normal air using an animal ventilator (Harvard Apparatus INSPIRA ASV 55-7058). Either kidney was then surgically exposed in a minimally invasive manner and kept moist with saline for the duration of the experiment. The mice were then placed under a LSM (Olympus FV1000). 2.2.Holder StabilizerIn order to minimize tissue motion, the organ of interest was stabilized using a custom-made mechanical holder (Fig. 1). The holder is composed of two stabilizing arms made of rigid aluminum alloy bent into a -shape, and a 150 micrometer thick coverslip [Fig. 1(b)] with a dimension along the maximum axis of 5 mm. The two arms and the coverslip thus constitute three-dimensional boundary constraints, which act to minimize tissue motion; the arms suppress horizontal movements while the coverslip restricts vertical movements. The use of a coverslip as an organ compressive solution is common in imaging studies. However, its sole use does not produce satisfying motion suppression. While the coverslip is capable of restricting vertical motion, it is unable to restrict the horizontal components of movement while at the same time does not guarantee reproducibility in position. Rather, vertical motion is converted into horizontal motion amplifying horizontal displacement. Fig. 1(a) The organ of interest is surgically exposed in a minimal invasive manner and the area to be imaged is softly compressed by the custom-made mechanical holder. LSM: laser scanning microscope, VN: animal ventilator. (b) The holder is composed of two holding arms made of rigid aluminum alloy, and a coverslip. Bar corresponds to 5 mm. (c) Compressive action of the mechanical holder on the imaged tissue. Arrows indicate the area of the tissue bounded by the aluminum arms and coverslip. The synchronization between acquisition and ventilation is achieved either by changing the microscope acquisition imaging parameters (image size, imaging speed) or by slightly adjusting the frequency of the ventilator ( from the mean normal rate of 134 BPM). Retrospective breathing-gated unsynchronized acquisition is obtained by recording both ventilation pressure signal and the line scan signal of the microscope. 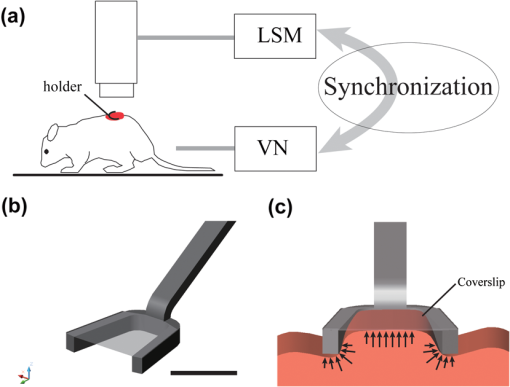 The stabilizing mechanical holder used in the present study is connected to a monolithic tri-stage base plate; this facilitates the holder’s orientation in space by allowing micro-adjustments and delicate positioning. 2.3.Image Stabilization Through SynchronizationThe use of the custom-made mechanical holder resulted in a significant suppression of respiratory-associated motion, namely the range of tissue motion was reduced from a few millimeters to several micrometers. While the reduction was substantial, the remaining tissue motion components were still large enough to cause distortions in the acquired images as well as unstable imaging sessions at high magnification. One possible solution to this problem could be to apply greater pressure to the tissue using other holders or a simple coverslip. Alternatively, the exposed organ could be placed face down in an inverted microscope, with the mouse body weight acting as a pressure stabilizer to suppress tissue motion. The first option is generally not appropriate since the high pressure exerted on the tissues would not only have a negative effect on their physiological function but would also cause harmful damage to the subject. While the second option is a viable possibility, it is not always feasible for organs that cannot be easily exteriorized such as the arteries, carotids, liver, lymph nodes, etc. Furthermore, due to the inverted configuration of the system, sophisticated positioning of the tissue cannot be achieved. In alternative to these options, we decided to synchronize image acquisition with the mouse’s breathing pattern. Because normal breathing is not precisely periodic either in time and/or displacement, we used a mechanical ventilator to regularize respiration, rendering it predictable and reproducible. Final image synchronization was thus achieved by either adjusting the image acquisition time and/or the respiratory rate. With a proper choice of these parameters, images acquired at different time points will be highly correlated with each other thanks to the reduced range of motion and reproducibility in position. This idea is illustrated in Fig. 2(a). Fig. 2Scheme of principle for synchronous and retrospective breathing-gated imaging acquisition. represents the displacement of the imaged organ along the vertical direction as a function of time; for simplicity is modeled as a sinusoidal wave of amplitude A and period . During the acquisition time each image is sampled at a frequency which is equal to the inverse of the dwell time (i.e. pixel integration time), with each pulse (green dotted lines) corresponding to a single image pixel. is the time interval between two consecutive images. (a) If the sample motion will be synchronized with the image acquisition. Consecutive images and will then be equal and will sample the imaged tissue always at the same positions (red curve). (b) If image acquisition and phantom motion are not synchronized, consecutive images and will then sample the phantom at different depths (red curve). This leads to a sequence where all images are different from each other. (c) Having knowledge of the motion function , it is possible to consider a specific time gating window triggered on a particular phase of the respiratory function. For the case here illustrated we consider a window centered around time points with zero derivative in (red curve). Images will then be sampled in correspondence to these areas. (d) By collecting several images () it is possible to extract from each one a portion of image (“patch”) which corresponds to the same temporal window . Finally, combining different patches it is possible to reconstruct an image where each point corresponds to the same height in within the imaged volume, i.e. the image is acquired as in a condition of absence of motion. 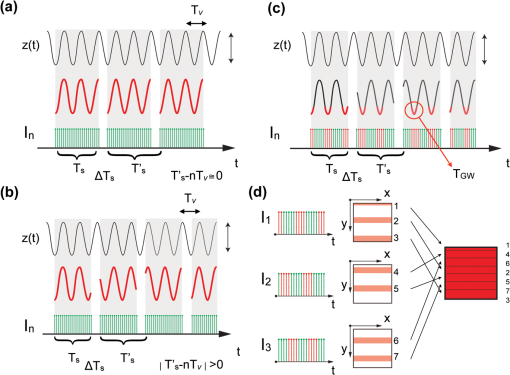 During image acquisition, the excitation scanning point draws a continuous raster path which is a periodic function of time: where is the time between two consecutive images, is the image acquisition time, and is the time interval between the end and the beginning of two acquisitions. The imaged sample instead moves with a generic periodic motion controlled by the mechanical ventilator and given by: where and correspond to the minimum and maximum ventilation period, respectively. Images acquired at different time points will then be equal [Fig. 2(a)] if (; synchronized acquisition) while they will be different for all other cases, i.e. unsynchronized acquisition [Fig. 2(b)].For a commercial laser scanning microscope, the image acquisition time (and ) can only assume a reduced number of values () according to the selected image size and/or scanning speed. Table 1 lists different possible settings for images acquired at a speed of 2 μs per pixel, using an Olympus FV1000 confocal microscope. Table 1Image acquisition time for Olympus FV-1000
Acquisition of images, using a frame rate of 139.9 frames per minute (FPM), would be a good option for synchronization since the mouse could be ventilated at the same rate of 140 breaths per minute (BPM). Alternatively, other combinations could be likewise selected for synchronized imaging. For example, pixel images, acquired at a frame rate of 71.8 FPM, could be used with a ventilation rate of 144 BPM. For this case, the respiration rate would be set at a frequency that is double the image acquisition rate (), taking into account the normal breathing cycle of the mouse. 2.4.Image Reconstruction from Retrospective Breathing-Gated Unsynchronized AcquisitionA synchronized acquisition strategy enables reproducible observations of the same area [Fig. 2(a)]. Unfortunately, the points belonging to the microscope imaging plane lie all at different depths within the observed organ due to the presence of the temporal motion component induced by the animal’s breathing (Fig. 3). Fig. 3Laboratory (black) and sample (red) reference frames are used to describe the position of a point located within a sample, which moves along the vertical direction with a sinusoidal path described by . The function describes the scanning path of the raster beam in the laboratory frame . (b) If an observer is positioned in the laboratory frame , the scanning path lays on a horizontal plane while the sample will appear moving in a sinusoidal fashion. (c) If an observer is positioned within the sample (reference frame ), the scanning path sampling the phantom will appear as laying on a curved surface . Images of moving objects will then represent points belonging to actual curved surfaces within the sample. 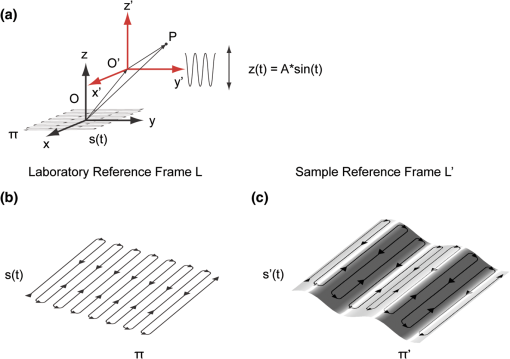 In the time during which an image is scanned, the excitation scanning point will move, with respect to the laboratory frame , on a horizontal imaging plane along a trajectory [Fig. 3(b)]. When considering instead a reference frame with respect to the observed organ, the imaging surface will be modulated in time by the breathing frequency [Fig. 3(c)]. The trajectory in the sample frame can therefore be easily obtained with a simple change of base: where the motion function corresponds to the organ displacement along the vertical direction, and is the vertical coordinate of . As a result, the acquired image will not coincide with any horizontal plane intersecting the organ but will instead resemble a curved surface and therefore will be intrinsically affected by severe distortions. This is in contrast to what happens in retrospective or prospective gated CT, where all pixels within an image are simultaneously acquired. In fact, for laser scanning microscopy, typical acquisition times are in the order of the respiratory cycle , (, ) with each pixel sequentially acquired at different times [Fig. 2(a)–2(c)] during .Breathing motion-induced artifacts could be eliminated by stopping the ventilator []; the imaging scanning point will then move on a surface and images will be representative of horizontal imaging planes within the tissue. Because breathing suppression is not feasible for extensive in vivo imaging sessions, we decided to use a retrospective breathing-gated approach to trigger selective image extraction at specific time points within the respiratory cycle. We therefore collected a series of images [Fig. 2(c)], and from each one we considered all pixels that had been sampled within a specific time gating window . By choosing appropriately both acquisition and motion periods (unsynchronized acquisition), this temporal portion area of the original image (“patch”) will span over time the entire objective’s field of view [Fig. 2(d)]. Using direct knowledge of the motion function and choosing a gating window centered around time points with the same vertical coordinates, it would be possible in principle to combine all the patches selectively extracted from each image and to reconstruct an image which is representative of an actual horizontal optical sectioning plane within the organ [Fig. 2(d)]. In Fig. 2(c) as an example, we chose a temporal window centered around time points with zero derivative in (red curve segments). But because is not always easily obtainable, we have to rely instead on the directly measured ventilation pressure waveform . With this information, optimal reconstructions can then be obtained by centering the gating window on specific intervals of the respiratory cycle located towards the end of the inspiration or expiration phases. While both a prospective and a retrospective method allow for a steady-state imaging, we here chose to implement a retrospective gating modality because not all commercial microscopes offer the option to be directly gated on the acquisition. To validate our approach we have prepared a phantom of agarose embedded with fluorescent beads. The phantom was kept in motion along the vertical direction in order to mimic a mouse’s breathing pattern; for simplicity we consider here a sinusoidal periodic motion with period and amplitude A. Each acquired image will correspond to a specific optical sectioning plane within the sample. When no motion is present [Fig. 4(a)], the imaging plane corresponds to a flat horizontal plane in both the sample and the laboratory frame () and images taken at different times are coincident (). When instead a vertical motion component is present, the imaging plane in the sample frame will appear as a curved surface Figure 4(b) with a characteristic periodicity related to ; here different pixels in the acquired image represent points with different depths within the agarose sample. If motion and acquisition are synchronized, all the curved surfaces corresponding to different images will overlap [Fig. 4(b)] and all acquired images will be equal to each other (). Fig. 4A phantom of agarose, with embedded fluorescent beads, is imaged with a laser scanning confocal microscope. In order to mimic vertical periodic motion, the phantom is vertically displaced with a sinusoidal function with a frequency of 2.3 Hz. An image sequence is recorded and two consecutive images , (taken at different time points and ) are shown. For different acquisition condition we show both a 3D representation of the phantom combined with the optical sectioning imaging planes, and the two images , . (a) The phantom is imaged with no vertical motion (i.e. ). The imaging plane consists of a horizontal plane () and thus the resulting imaging sequence is stationary. Images and are equal. (b) The phantom is subjected to a periodic vertical motion component (); this results in a distortion of the imaging plane along the axis giving rise to a curved surface . Here both image acquisition and motion are synchronized in time. The imaging plane forms a sinusoidal plane but images remain identical across different time points within the same image sequence. As a result, the image sequence appears stationary. (c) If image acquisition and motion are not synchronized, images taken at different time points and sample different areas of the phantom, therefore images within the same sequence appear different. (d) Scheme of principle for obtaining a reconstructed image starting from a sequence of images acquired using a retrospective breathing-gated imaging modality with selective image extraction gated to the temporal window . 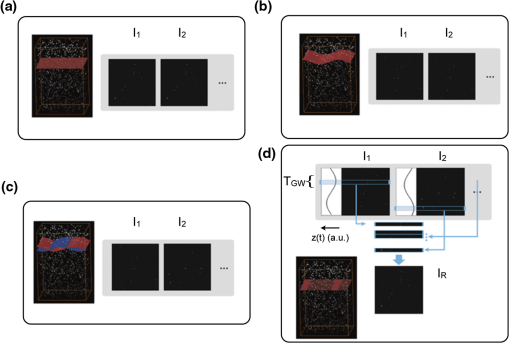 If motion and acquisition are not synchronized, the imaging planes will gradually sample the phantom, over time, in the whole volume delimited by the motion’s amplitude A and the objective’s field of view [Fig. 4(c)]. By collecting a sufficient number of images (i.e. sampling surfaces), we can reconstruct both a single horizontal plane within the sample [Fig. 4(d)] as well as a three dimensional rendering of the sampled volume. In a similar manner, we were able to obtain in vivo motion-free and artifact-free images of a mouse organ by using direct knowledge of the ventilation pressure waveform . The image acquisition time and ventilation rate were adjusted to be slightly out of synchrony. In addition, both the ventilation pressure from the animal ventilator and the line scanning signals of the microscope were recorded during image acquisition to facilitate reconstruction. To avoid abrupt changes in pressure and to help produce stabilized images, a fixed time gating window , corresponding to a particular interval of the respiration cycle near the end of inspiration, was selected from the recorded pressure signal. Excellent reconstructed images could be also obtained by centering the window near the end of expiration. Figure 5 shows the acquired images together with their corresponding lung pressure curves; the red boxes represent the time gating window . In a similar fashion as illustrated in Fig. 2(d), data collected over time within these temporal windows were then fused together to produce a reconstructed motion-free image which is representative of an actual horizontal optical sectioning plane within the organ (Fig. 5). Fig. 5Scheme of principle for in vivo retrospective breathing-gated imaging. The procedure is similar to the one used for the phantom imaging. Image acquisition and ventilation are kept unsynchronized, and both ventilation pressure signal and the line scan signal of the microscope are recorded. A gating window centered on specific intervals of constant pressure within the respiratory cycle and located towards the end of the inspiration or expiration phases is chosen. Data from different acquired images that fall within the temporal window are collected. These data are then merged together to reconstruct artifact-free images which are representative of actual horizontal optical sectioning plane within the imaged organ. 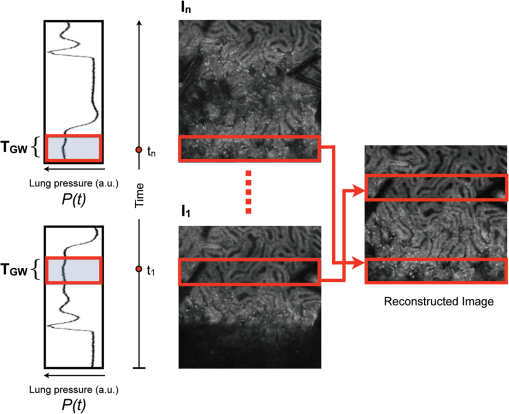 3.In Vivo ResultsIn vivo image sequences of a kidney in a ubiquitin/green fluorescent protein (GFP) expressing mouse were acquired. In Fig. 6(a), an image sequence obtained using a conventional set-up without a stabilizer is shown. Here, due to tissue movement, different areas of the kidney are observed to enter and leave the focal plane of the imaging objective. When our custom-made mechanical holder is used with the described set-up and no synchronization between motion and acquisition is present [Fig. 2(b)], the motion components are greatly reduced [Fig. 6(b)], increasing in-focus areas by approximately three-fold. Residual tissue movement however, albeit minimal, still gave rise to image artifacts due to the fact that different parts of the kidney are travelling at different speeds during image acquisition. Here images were acquired at a speed of 139.9 FPM while the ventilation was set at a speed of 134 BPM. Fig. 6(a) An image sequence acquired using a conventional set-up without any stabilization. Different areas appear in the focal plane of the image due to movement of the tissue. b) An image sequence acquired using only the custom-made mechanical holder. Images appear more stable compared to (a), but residual movements are still present and result in image artifacts. Arrows indicate different areas among images. c) An image sequence acquired using the combined approach comprising both the custom-made mechanical holder and the gating synchronization. Images obtained are highly stabilized. (d) and (e) Plots depicting the planar position of a fluorescent bead injected intravenously before imaging: (d) without or (e) with respiratory gating synchronization. One pixel corresponds to 2 micrometers. Measurements of the bead’s movement using the conventional set-up were not possible as the large motion component of the tissue compromised the bead’s visibility over time. 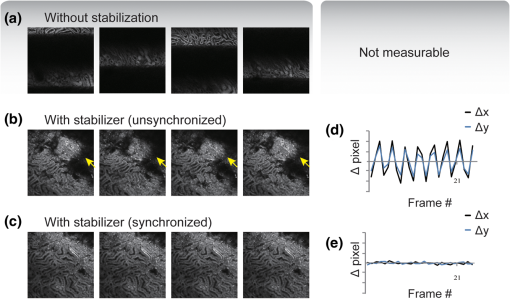 When motion and acquisition are instead synchronized, in combination with the mechanical holder’s presence, the acquired images [Fig. 6(c)] appeared highly stabilized and residual respiratory artifacts were ameliorated. In this case images were acquired at a speed of 139.9 FPM while ventilation was set at a lower speed of 140 BPM. To subsequently quantify the improvement attained using our combined approach, we injected fluorescent beads (of 5 μm diameter) into the tail vein of the mice. Within minutes, several of the beads became trapped in the capillaries. These were then used as fiducials to better characterize the degree of image stabilization by measuring the beads’ positions as a function of time [Fig. 6(d) and 6(e)]. An increase of stabilization was observed from 2.55 pixels (5.07 micrometers) when only the holder is implemented, to 0.24 pixels (0.48 micrometers) using the synchronized imaging modality—in other words, a improvement in image stabilization [Fig. 6(d) and 6(e)]. Despite the lower reduction in motion stabilization and the presence of distortions within the acquired images (i.e. images represent curved surfaces and not horizontal planes), synchronized acquisition offers the advantage of enabling the recording over time of dynamic events such as cell migration or vessel perfusion. Normally this is not possible in a retrospective approach where several images need to be collected upfront. Figure 7 depicts images of a live mouse kidney obtained in synchronized mode in combination with our stabilizer; green represents the GFP signal from GFP-expressing renal cells and red represents the fluorescence signals from Rhodamine B previously injected via tail vein; merged images are shown in the far right panels. Even at high resolution, images appear stable and without artifacts. Fig. 7Images of a live mouse kidney; green represents the GFP fluorescence signal from GFP-expressing kidney cells while red represents the fluorescence signals of Rhodamine B, used here as a blood pool contrast agent. Fused images are shown in the far right panels. All images were acquired using a water immersion objective (Olympus, XLPlan N NA 1.05). Image acquisition and ventilation are here kept in synchronization. The optical sectioning imaging plane is a curved surface but consecutive images are highly correlated allowing for fast dynamic imaging. (a) Images acquired with a objective. (b) Images acquired with a objective. 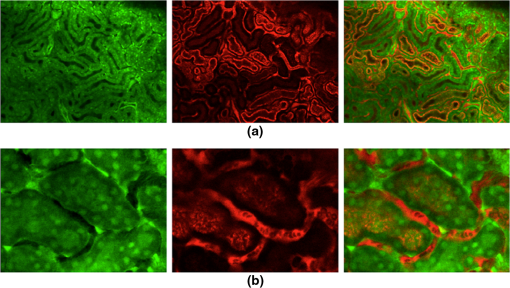 We then performed retrospective selective image extraction gated near the end of the mouse inspiration phase. Figure 8(a) shows a reconstructed image of a kidney in a breathing mouse, obtained by combining all patches selectively extracted from a sequence of images . The reconstructed image is representative of an actual horizontal optical sectioning plane within the kidney. Here, images were acquired at a speed of 139.9 FPM while ventilation was set at a lower speed of 134 BPM. was equal to 48 ms and 25 images were collected for final reconstruction. Fig. 8Comparison between a reconstructed image using the combined approach comprising both the custom-made mechanical holder and the retrospective breathing-gated acquisition with selective image extraction (a) and an image acquired using the ‘stop-ventilation’ technique (b). The high degree of correlation between the two images indicates the goodness of our method and the lack of artifacts in the final reconstructed images. All images were acquired using a water immersion objective (Olympus, XLPlan N NA 1.05). (c) By controlling the vertical position of the imaging objective and by imaging at different depths over time it is possible using our combined approach to obtain three dimensional reconstructions of entire volumes within moving organs. (d) and (e) Images representing horizontal imaging planes within the 3D stack. 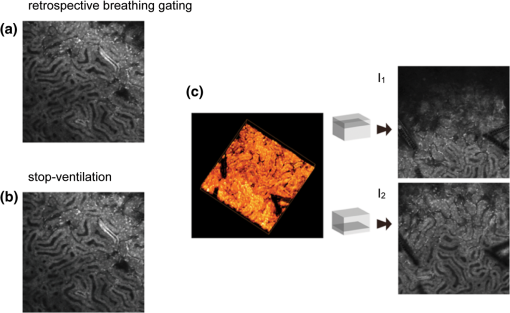 Next, to verify the accuracy of the reconstructions and to prove that no artifacts had been introduced while combining the different patches, we compared reconstructed images to images acquired using a ‘stop-ventilation’ technique,2 wherein breathing motion is suspended for 30 s. The kidney was then imaged at varying heights by adjusting the vertical position of the objective in order to find the plane corresponding to the one previously reconstructed during ventilation. From this comparison, we found a high correlation (Pearson’s coefficient ) between the reconstructed image [Fig. 8(a)] and the one acquired via the ‘stop-ventilation‘ method [Fig. 8(b)]. This thus served to verify that our approach was indeed successful in producing accurate images without any reconstruction induced artifacts. Finally, using the described combined imaging modality we were able to achieve in vivo artifact-free reconstructions of three-dimensional volumes within the moving kidney. By simply acquiring a sequence of images and varying the vertical position of the imaging objective over time, reconstructions of actual horizontal planes at different heights were obtained and then stacked over each other [Fig. 8(c)]. 4.ConclusionsHerein, we propose a practical motion stabilizing technique, using a custom-made mechanical holder, for use with intravital confocal laser scanning microscopy. The proposed method has distinct advantages over conventional methods; namely 1. it can be easily applied to most commercial confocal and multiphoton microscopes as are used by most biological imaging laboratories without the need for modifying hardware, and 2. it provides motion-stabilized image sequences without imparting a negative effect on the subject. In vivo experiments demonstrated that the range of motion within the image sequences using the technique decreased from several millimeters to micrometers. Moreover, we demonstrated that by using ventilation pressure signals, motion-free and artifact-free images, in which the pixels correlate with particular depths within the imaged organ, could be achieved. This technique could be used to achieve motion-free images of tissues that are significantly affected by movement artifacts such as the liver, arteries, carotids etc. AcknowledgmentsThis project was funded in part by Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (under Contract No. HHSN268201000044C), and from the Institute of Biomedical Engineering (under R01EB006432). ReferencesA. Bullen,
“Microscopic imaging techniques for drug discovery,”
Nat. Rev. Drug Discovery, 7
(1), 54
–67
(2008). http://dx.doi.org/10.1038/nrd2446 NRDDAG 1474-1776 Google Scholar
E. C. Rothsteinet al.,
“Multi-photon excitation microscopy in intact animals,”
J. Microsc., 222
(1), 58
–64
(2006). http://dx.doi.org/10.1111/jmi.2006.222.issue-1 JMICAR 0022-2720 Google Scholar
M. R. Looneyet al.,
“Stabilized imaging of immune surveillance in the mouse lung,”
Nat. Methods, 8
(1), 91
–96
(2011). http://dx.doi.org/10.1038/nmeth.1543 1548-7091 Google Scholar
S. Leeet al.,
“Image Stabilization for in vivo microscopy by high-speed visual feedback control,”
IEEE Trans. Robot., 24
(1), 45
–54
(2008). http://dx.doi.org/10.1109/TRO.2007.914847 IRAUEZ 1042-296X Google Scholar
S. Laffrayet al.,
“Adaptive movement compensation for in vivo imaging of fast cellular dynamics within a moving tissue,”
PLoS One, 6
(5), e19928
(2011). http://dx.doi.org/10.1371/journal.pone.0019928 1932-6203 Google Scholar
D. S. GreenbergA. R. HouwelingJ. N. Kerr,
“Population imaging of ongoing neuronal activity in the visual cortex of awake rats,”
Nat. Neurosci., 11
(7), 749
–751
(2008). http://dx.doi.org/10.1038/nn.2140 NANEFN 1097-6256 Google Scholar
D. A. Dombecket al.,
“Imaging large-scale neural activity with cellular resolution in awake, mobile mice,”
Neuron, 56
(1), 43
–57
(2007). http://dx.doi.org/10.1016/j.neuron.2007.08.003 NERNET 0896-6273 Google Scholar
I. S. Veilleuxet al.,
“In vivo cell tracking with video rate multimodality laser scanning microscopy,”
IEEE J. Sel. Topics Quantum Electron., 14
(1), 10
–18
(2008). http://dx.doi.org/10.1109/JSTQE.2007.912751 IJSQEN 1077-260X Google Scholar
R. T. A. Megenset al.,
“In vivo high-resolution structural imaging of large arteries in small rodents using two-photon laser scanning microscopy,”
J. Biomed. Opt., 15
(1), 011108
(2010). http://dx.doi.org/10.1117/1.3281672 JBOPFO 1083-3668 Google Scholar
M. Lieblinget al.,
“Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences,”
J. Biomed. Opt., 10
(5), 054001
(2005). http://dx.doi.org/10.1117/1.2061567 JBOPFO 1083-3668 Google Scholar
S. Giouxet al.,
“Motion-gated acquisition for in vivo optical imaging,”
J. Biomed. Opt., 14
(6), 064038
(2009). http://dx.doi.org/10.1117/1.3275473 JBOPFO 1083-3668 Google Scholar
N. A. Scottet al.,
“Systematic review of beating heart surgery with the Octopus Tissue Stabilizer,”
Eur. J. Cardiothorac. Surg., 21
(5), 804
–817
(2002). Google Scholar
|

