|
|
1.IntroductionSkin optics is a research area rich in applications involving diagnosing, characterizing, and understanding the properties of tissues and organs. For over three decades, the area has attracted many research groups that have targeted a host of different applications. Several studies have attempted the task of developing tools to estimate the biological parameters that make up the layers of skin. This is an important task because quantitative knowledge of these parameters can be invaluable in applications such as medical diagnostics, wound-care, drug-delivery, and skin aging, amongst others. Many studies have cited1–3 that skin parameters can be used in a longitudinal study to trace the growth and spread of skin cancers. They can also be used to understand the pathophysiology of malignant tumors. Therefore, technologies that can estimate skin parameters can help in diagnosing cancers such as melanoma, which results in approximately six deaths every hour worldwide.4 Such technologies can be especially effective because the most common diagnostic for diseases such as melanoma remains a nonobjective visual examination by a health care professional.5 Traditionally used for remote sensing, hyperspectral imaging has recently received increased attention for its ability to automatically detect and classify anomalous areas in a wide variety of biological materials, including the human skin.6 A hyperspectral imaging system is able to measure specific spectral signatures based on the pigmentation and the color of human skin in the visible (UV-VIS) through the short-wave-infrared (SWIR) regions of the electromagnetic spectrum.6 In this study, we use hyperspectral radiometric measurements in order to detect the optical properties of biological materials by reporting their reflectance spectra. This reflectance signature can be used to develop a model that forward maps the skin parameters to their hyperspectral signature. This is the “forward model” discussed in this article—computing the reflectance spectra of a skin sample based on the underlying biological parameters. The novel aspect of this article is solving the inverse problem–using machine learning regression to estimate the skin parameters from the hyperspectral signature. This article is organized as follows. Section 2 reviews some prior work in skin optics. Section 3 provides an overview of the physics-based forward model. Section 4 reviews the derivation of the machine learning based inverse model. Section 5 includes the experimental methods and results. Section 6 discusses the results and validations. Finally, Sec. 7 includes our concluding remarks. Preliminary portions of this work are reported in conference proceedings.7–9 2.Review of Prior WorkOne potential taxonomy of research in skin optics is as follows: first, studies focused on developing robust biophysical models of human skin; second, studies focused on developing methods to estimate the biological components of skin; and third, studies focused on using computational models, coupled with imaging modalities, for medical diagnostics. In this section, we review some of these studies, and also highlight our own contribution. The first group of studies deals with developing models of how light interacts with human skin. The goal of these studies is to create models that can represent skin and all of its optical and biochemical properties using simply the measured spectral power distribution. This power distribution is given by the measured reflectance and transmittance.10 One of the earliest such models was developed by Kubelka and Munk who related the reflectance spectra of paint to its absorption and scattering coefficients.6,11 The Kubelka-Munk (K-M) theory based model uses the absorption and the scattering coefficients as inputs for the energy transport equations to describe the transfer of radiation in scattering media (such as human skin). The K-M model is also referred to as the “Flux Model” because it incorporates two fluxes (diffuse upwards and downwards fluxes). Since the original work by Kubelka and Munk, the model has been improved and optimized by several groups. Van Germet and Star12 expanded the K-M model to account for tissue scattering behavior. Tuchin et al.13 and Yoon14 built on this work and incorporated four and seven fluxes, respectively, to account for the radiation scattering. Meglinski and Matcher15 developed a hybrid K-M model that uses the Fresnel equations to speed up the model output. Nunez6 modified this model by incorporating optical parameters coupled with in vivo and ex vivo measurements. They report better performance for their model compared with other K-M based models.6 Therefore, in this article, we employ their hybrid K-M model. Several groups have also developed variants of the K-M model such as the diffusion theory model, which uses the Boltzmann photon transport equation, the absorption and scattering coefficients, and a phase function.16 Other variants include the radiative transport model,17,18 and the Monte Carlo methods model,19 however these are typically used for laser applications. In addition, Baranoski and Krishnaswamy10 comment that for these models, the comparisons between modeled and measured data are seldom provided. Nonetheless, these models have been used by several groups20 for a host of biomedical applications. In contrast to the K-M variant models, Hanrahan and Kreuger21 (expanded by So-Ling and Li22) developed a scattering model, i.e., the H-K model, which models skin as two-layers, epidermis and dermis. Finally, Stam et al. developed the discrete-ordinate model, i.e., the D-O model, which has been used to simulate the scattering behavior of human skin.23 The D-O model treats skin as a single layer with homogeneous optical properties and index of refraction. While each model has advantages and disadvantages, the K-M based models offer the greatest compromise between computational efficiency and accuracy.10 The second group of studies focuses on developing methods to estimate human skin parameters. In particular, several groups have used variants of the K-M model in order to estimate skin parameters. Our study best fits into this category. The following studies are the closest to ours, in terms of methodology and validation. Cotton24 and Cotton and Claridge25 employ a K-M theory based model to estimate the melanin concentration in skin. They have also developed26,27 an inverse model based on finding a set of optimal image filters to minimize the error between a mapping from color images to skin parameters. Doi and Tominaga28 have also used a K-M model, coupled with a least squares method, to fit measured and estimated reflectance spectra in order to estimate skin pigments (e.g., melanin, etc.). Yudovsky and Pilon29,30 developed a semi-empirical model for diffuse reflectance of two-layered media by approximating the solution of the Radiative Transfer Equation (RTE). They then estimate a set of skin parameters using this optical two-layer model and an inverse method based on least squares minimization. We compare our methods, results, and validation to these studies in Sec. 6. In addition, there also exists work by Anderson and Parrish31 and Wan et al.32 who used the K-M model to estimate the reflectance of dermal and epidermal tissues in vivo (but the physical skin parameters). Tsumura et al.33–35 estimated skin chromophores using Independent Component Analysis, and by matching simulated reflectances to the estimated reflectances at each pixel of a multispectral image. Claridge et al.36 created a model of skin and tissue coloration by finding the spectral composition of light remitted from skin parameters. They use this model to find a mapping between color images and pigmented skin lesions. Some groups, such as Alander, Kaartinen, Leonardi, Zerubia, et al.,37–41 have also used hyperspectral and multispectral imaging, often coupled with K-M model variants and/or inverse methods, to estimate skin chromophore concentrations and classify skin pigmentation. The last group of studies apply computational models and medical imaging modalities to medical diagnostic applications. In particular, these studies apply the methods from the previous two groups of studies to a clinical setting. For example, Yudovsky et al.42 collected hyperspectral oximetry data from 54 in vivo subjects with some degree of foot ulcers. They then used their algorithms to classify the subjects into two groups: one, whose ulcers healed within 24 weeks, and the second, whose ulcers did not heal within 24 weeks. They based their classification on the estimated concentrations of oxyhemoglobin, de-oxyhemoglobin, and oxygen saturation. Similar studies by other groups include classifying skin lesions from images,43 investigating skin alterations in diabetes patients,44 and assessing hemodynamic changes in skin, post-burn,45 amongst several others. The studies presented here by no means constitute a complete review of the skin optics research area. However, they do highlight the rich history of the area, and also help indicate the novel features of our work. These features include:
3.Physics-Based Forward Modeling3.1.Human Skin ModelThe model of human skin presented here is based on the models derived by Meglinski and Matcher15 and later modified by Nunez6 Skin is treated as an -layered Lambertian material, which allows for uniform bidirectional reflection. The 10 layers are defined as follows: Layers 1 through 5 represent the strata of human skin; these layers are: stratum corneum, stratum lucidum, stratum granulosum, stratum spinosum, and stratum basale. Layers 6 through 9 are the dermises; these layers are: papillary dermis, upper blood net dermis, reticular dermis, and deep blood net dermis. Layer 10 is the subcutaneous tissue layer, and it is assumed to be infinitely thick, allowing the model to ignore the transmittance through it, as it is zero for a layer with infinite thickness. Skin is composed of seven biological parameters. In this work, we estimate five such parameters. These parameters are: melanosome concentration (), collagen concentration (), -saturation (), and subcutaneous reflectance (). The physiological ranges for each estimated parameter are detailed in Table 1. The last two parameters (layer thickness, , and water volume ) are assumed known and hence not estimated. Table 1Biological parameter detailed descriptions and physiological ranges.
In order to make the model tractable, the following underlying assumptions (as described by Nunez6) are made. First, each layer is assumed to have similar optical properties, and homogeneous absorption and scattering coefficients. This means that the concentration of each parameter is the same for all the layers. The parameter is an internal model parameter that doesn’t have any physiological meaning. Therefore, it is omitted from further analysis. Therefore, we are estimating four parameters in this study (omitting ). Next, each layer has a particular thickness, and a water percentage. These thicknesses, and water percentages are assumed known and not estimated. They are kept constant for each layer based on work by Meglinski and Matcher;15 these are tabulated in Table 1. Finally, blood is assumed to be uniformly distributed in the dermis layers (rather than in differing concentrations for each layer) and zero in the strata. While some of these constraints may not be consistent with real human skin, Nunez et al. demonstrate minimal modeling error despite these underlying simplifications.6 3.2.Forward ModelThe proposed method is based on a physics-based forward model that describes the reflectance spectra of human skin based on physiological optical parameters that make up its layers. The forward model describes a method of modeling the reflectance spectra of each layer based on the knowledge of each layer’s thickness and the optical properties of its constituent components.6,48 In general, the forward mapping can be described as follows: where is a vector containing the skin parameters (see Table 1), and represents the corresponding hyperspectral signature vector.3.3.Kubelka-Munk TheoryThe relationship described by Eq. (1) is based on a set of analytical models6,48 that describe the transmission, , and reflection, , of light at a specific wavelength in a layer of (biological) material, where denotes the layer number. The reflection and transmission are computed using the K-M equations, given by: with the following parameters tied to the absorption and scattering properties of the biological materials: where denotes the thickness of layer , and is equal to . The coefficients, and are based on the absorption, , and the scattering, , coefficients and are given by: The coefficient is computed separately for each strata (layers 1 through 5) and dermis (layers 6 through 9), and is based on the components of as seen by: where , , and , are the absorption profiles of the biological materials (melanosome, collagen, water) contained in based on empirically derived values tabulated in Ref. 6. The coefficients , , , and are the absorption profiles of betacarotene, oxygenated hemoglobin, de-oxygenated hemoglobin, and bilirubin, whose absorption profiles are also included in Ref. 6.3.4.Light Transport ModelIn addition to using the K-M equations, we also use the Fresnel equation, as detailed by Nunez6 Since human skin has uniform bidirectional reflection, Nunez et al. assume that light incident on the skin surface is always normal to the surface. This allows us to use the Fresnel equation to describe the amount of reflection that is normal to the interface separating the skin from air. The equation is based on the tabulated indices of refraction for air and the stratum corneum (skin layer 1), given by , and , respectively. The Fresnel reflection FR is then given by: The reflectance path of light is modeled in the following fashion. For every layer, light can take one of four paths: it’s either (1) absorbed, (2) scattered out of the top of the layer, (3) scattered out of the bottom of the layer, or (4) doesn’t scatter, and continues along its path. Therefore, the reflectance and transmittance between any two interfaces (layers) are going to be infinite sums dependent on what path light takes between those two interfaces.In addition, light being reflected off the surface of the skin is actually made up of light coming from each path leaving a skin layer, as well as the Fresnel reflection. In other words, the total fraction of light leaving each layer is the product of: (i) the Fresnel transmittance, (ii) transmittance of all the layers it had to go through in order to each layer , (iii) the reflectance of the layer (the model assumes an infinitely thick bottom layer, and therefore has pure reflectance), (iv) the transmittance of all the layers it must once again traverse in order to reach the top, and finally (v) the Fresnel transmittance. Note that the transmittance from layer 1 to layer is the same as the journey back from layer to layer 1. Using this methodology, the the total reflectance and transmittance between any two interfaces is the sum of all reflectance paths (see steps 1 through 4 above) and the transmittance and reflectance it accrues from its journey [see steps (i) through (v) above]. This is given by: where , and , are the total reflectance and transmittance between interface and . The other variables are computed using Eqs. (2)–(5) from above. A graphical version of this, expanded from Ref. 6, is presented in Fig. 1.Fig. 1A graphical representation of Eq. (7); the light transport model for possible paths of light between any two interfaces (layers). 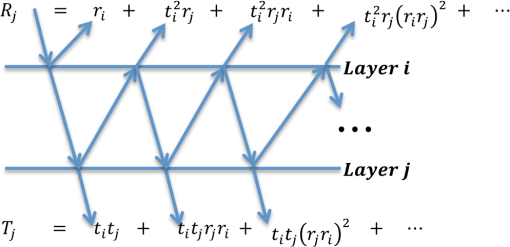 The final step is to iterate the quantities in Eq. (7) for all optical layers 1 through . This is analogous to the procedure presented in Fig. 1 and Eq. (7), except adapted for all layers of the skin, until the overall reflectance (and transmittance ) is computed. The closed form solution is given by: Using this methodology, the reflectance spectra of human skin, given by , can be generated in the UV-VIS through the SWIR regions of the electromagnetic spectrum based on the physiological parameters . 4.Machine Learning-Based Inverse ModelingIn Sec. 3, we described a physics-based model that maps the physiological parameters of skin to its observed reflectance spectrum. This section presents two methods to compute the inverse map, answering the question: “given a reflectance spectrum of human skin, what are its underlying physiological parameters?” This problem can be expressed mathematically as follows: which is the inverse of Eq. (1). We adopt two machine learning regression algorithms to estimate the inverse mapping function : (a) support vector regression (SVR)49,50 and (b) -nearest neighbors based regression (-NN).51The use of regression is motivated by the availability of a large number of training spectra and their associated physiological parameters . It leverages the abundance of this data to yield an estimate of the parameters of with minimal error and low variance. In contrast to other studies to estimate skin parameters, we utilize machine learning methods because they provide the following benefits: Nonparametric: the functional form of is not known and it cannot be analytically computed. The learning paradigm is data-driven and does not require any a priori knowledge about the functional form of . Generalizable: these methods avoid over-fitting and hence offer good results to unseen data when compared to traditional methods. Kernels: these methods can exploit the kernel trick to use linear regression to accurately model nonlinear functions. Since one cannot assume linearity for , machine-learning regression can be a powerful tool for learning the nonlinear function that maps the physiological parameters to their observed reflectance spectrum. 4.1.Support Vector Machine Based RegressionFor regression, we first use SVR, a now established machine learning approach, with several open source implementations (e.g., Ref. 52) in C++ and Matlab. The remainder of this section gives a high level motivation of the SVR technique and its implementation. The full mathematical details are left as a reference to Vapnik et al.49 and Smola and Scholkopf.53 To solve the regression problem in a way that can be approached using SVR, we decouple the function , which maps a spectral vector to a skin biological parameter vector , into a set of five (one for each component of , excluding , and , which are known, and hence kept constant) scalar regression subproblems where: where is one of the scalar parameters (components of ) we wish to estimate (e.g., collagen level, melanosome level, etc.), and is the scalar regression function. Each of the scalar parameters are then individually estimated in the same fashion.Because the approach is data driven, we start by considering the training dataset that was generated using the forward model described in Sec. 3.3. This consists of parameters vectors and their associated spectral vectors . We consider the set of pairs including one spectral vector and one biological scalar parameter we wish to estimate: In the simple linear case, the function can take a functional form given by: where represents the linear dot product. The regression function approximates the output parameter as a weighted linear combination of the input spectral vector , dot product with a set of spectral vectors , with an added offset . The goal is to find the and that satisfy an optimality criteria. Consider the regression margin, i.e., the distance such that all the training data points lie within this distance margin of the regression function . The optimality criteria used in SVR is to minimize the margin. As an additional improvement, some of the points are allowed to violate the margin constraint via the use of an additional slackness term, thereby allowing the method to remain robust to possible outliers. This results in a soft margin optimality criteria which is solved via a constrained optimization technique using Lagrange multipliers. When one invokes the Karush-Kuhn-Tucker (KKT) conditions,54,55 a set of support vectors and associated weights emerge from the complementary slackness constraint; these are the weights and vectors used in Eq. (12).53,56,57 These support vectors are some of the original training spectral vectors . The procedure just described makes up the training phase of SVR. As noted earlier, since a linear approximation is not sufficient to describe our skin dataset, we use a nonlinear dot product, commonly called a kernel, and denoted by . Then has the following expression: In sum, the regression machine uses the following steps: (a) train the machine to obtain , , and . Then, given any input test spectral vector , form the regression by (b) first taking the dot product between and each support vector , (c) these are then weighted by the weights , and (d) a linear combination is taken, which is (e) finally offset by the constant . This is conveniently implemented in Matlab and numerous Open Source implementations such as LibSVM52 or OpenCV.58 In particular to Matlab and LibSVM, two functions, namely svmtrain and svmpredict, implement the above training step (a) and regression steps (b)–(e), respectively. Nonetheless, a more thorough review of the SVR method is included in the Appendix.4.2.-Nearest Neighbors–Based RegressionThis learning algorithm is used in order to classify objects based on a “majority vote” system. Much like the SVR approach, this algorithm starts with a database of input training sets, known as the feature space, as presented in Eq. (11). The -NN algorithm regresses on a new testing set based on the closest training examples it finds in the feature space. In this work, the feature space consists of approximately 300,000 reflectance spectra generated as per Eq. (1). The testing set consist of in vivo hyperspectral signatures that obey Eqs. (1) and (9). The goal is to compute from Eq. (9). As is typical for hyperspectral signatures, the -NN algorithm needs to compare the shape of each hyperspectral signature from the testing set to the signatures from the training set. This is done by computing the inner product between the two signatures, and can be written as the spectral angle give by: where is a computationally generated hyperspectral signature from the training set contained within the feature space, is the signature obtained in vivo that is being classified and denotes the Euclidean norm.Alternatively, experiments are also performed using two other versions of the -NN algorithm: first, the closest neighbor is found by computing the Euclidean distance between spectra and second, the training and testing datasets are first whitened, and then spectral angle is used to compute the nearest neighbors. A more mathematically rigorous treatment of -NN regression is included in literature.51,59 5.ExperimentsIn this work, the goal is to compute the inverse mapping , and hence create a model that can infer the underlying constitutive physiological parameters of human skin from hyperspectral signatures. The evaluations are done through two sets of experiments. The first set involves performance validation through synthetic experiments. The second set involves performance validation through in vivo experiments. In both sets, a skin reflectance model, which is generated as per Eqs. (1)–(8), is used. The skin reflectance model (this model will be labeled the training dataset for the remainder of this article) is generated as follows. Following the physics-based model described in Eq. (1), approximately exemplars of all seven parameters described by are uniformly sampled on a grid and distributed along their entire physiological domain. These ranges are detailed in Table 1. An equal number of samples are generated for each . The forward model is used to generate a dataset of hyperspectral signatures corresponding to each set of parameters . The water level and the dermal thickness are kept constant for each layer as detailed in Ref. 6. This dataset is denoted as: This dataset is used for both sets of experiments. 5.1.Synthetic ExperimentsIn the first set of experiments, synthetic data is used to test the accuracy of the inverse model . Therefore, a new testing synthetic dataset is generated by using the forward model and a set of biological parameters that were not contained within the training dataset. Each set of parameters was generated by randomly sampling along the parameters’ physiological range. If a parameter was found to be too close to one already contained within the training dataset, it was discarded and a new one was generated. This ensured that there was no overlap between the training and testing datasets. The training set and the newly instantiated testing set are then used to perform the synthetic experiments. The synthetic testing dataset is denoted as: Tuples from the training dataset (15), , are used to train the SVR. The SVR algorithm employed here is implemented using the NTU SVM Library49,52 in Matlab. This produces the trained SVR model and its associated support vectors. Then, , as well as the SVR model is used to estimate . In these experiments, the values of and are already known, and therefore, they serve as the ground truth values. However, by using the SVR and only , as per , the estimated value of , given by, , can be computed. The estimated values, , can then be compared with the ground truth values, , in order to compute an error associated with the inverse mapping .The same experiment is repeated using the three flavors of the -NN regression algorithm. For each , the -NN (in this case, the one neighbor in the training dataset that had the smallest spectral angle, , with each ) were found. The estimated parameters, , are the parameters corresponding to the nearest neighbor spectra, i.e., . Once again, since the ground truth values of are already available (given by ), an error associated with the regression can be computed. These average absolute errors (AAE), for all four algorithms, along with the standard deviations (Std. Dev.) associated with the estimated biological parameters are provided in Table 2. The AAEs is computed as follows: Table 2Average absolute errors (AAE) associated with biological parameter estimation for synthetic experiments.
5.2.In Vivo ExperimentsThe in vivo experiments were performed using a dataset obtained from in vivo hyperspectral imaging of 24 individuals of both genders and Caucasian, Asian, and African American ethnicities. The data was obtained at Johns Hopkins Hospital, Department of Dermatology, under protocols approved by the Institutional Review Board (IRB). All patients gave informed consent, and the data was collected uniformly. Hyperspectral signatures were obtained from each of the 24 individuals using the Analytical Spectral Devices, Inc. (Boulder, Colorado) FieldSpec 3 Portable Spectroradiometer. The spectroradiometer has a hand-held probe which was positioned at a perpendicular angle to the skin, with the enclosed lens at a height of 5 cm from the skin. The spectroradiometer has a lens diameter of 10 mm, a field of view of 25 deg, a 100 ms scanning time, and a built-in illumination source. The spectroradiometer has two detectors, one containing a 512-element Si photodiode array (for imaging up to 1000 nm) and the other detector contains two graded index InGaAs photodiodes (for imaging beyond 1000 nm). The instrument is calibrated using a panel whose reflectance is known; the amount of light captured by the instrument is correlated with the reflectance of the panel for each wavelength. The instrument is recalibrated after each measurement. The spectra was obtained from 450 to 1800 nm blue with a 1 nm step size (bandwidth). The in vivo dataset obtained from this IRB is denoted as: This dataset was compiled by in vivo hyperspectral imaging of 24 individuals. Approximately ten hyperspectral signatures were collected from each individual, from five anatomical locations on their bodies (two signatures from each location), to sum to a grand total of signatures in the in vivo dataset. These five locations include: the back, the palm, the cheek, the dorsal forearm (DF) and the upper inner arm (UIA).The major difference between the in vivo experiments and the synthetic experiments is the ground truth. In the synthetic experiments, the estimated parameters are compared to the ground truth parameters , in order to assess the performance of the algorithms. The ground truth targets are not available for the in vivo dataset. However, we still perform performance validations for the in vivo dataset. The experiments using SVR and -NN are repeated in the same manner as described in Sec. 5.1. The estimated parameters are summarized in Table 3 and Fig. 2 as a function of anatomical location, and ethnicity. We analyze these results using physiological precepts in Sec. 5 of this article. In an effort to analyze the performance of the algorithms, we also report error bounds between the estimated and the ground truth spectra. In other words, Table 4 reports the spectral angle, the root mean square error (RMSE), and the standard deviation between the in vivo measured spectra and the -NN estimated spectra for each of the 24 patients. In Table 5 we build on this analysis, and report the spectral angle error as a function of anatomical location for each of the 24 patients. Finally, Fig. 3 shows examples of in vivo measured hyperspectral signatures plotted along with signatures estimated using the -NN algorithm. Table 3Estimated skin parameters (percentage by volume) as a function of ethnicity (Caucasian, Asian, African American) and anatomical location [dorsal forearm (DF), upper inner arm (UIA), back, cheek, palm].
Fig. 2Bar graph representations of the estimated (a) melanosome concentration, (b) collagen concentration, (c) oxygen saturation concentration, and (d) blood volume concentration, as a function of both ethnicity and anatomical location. 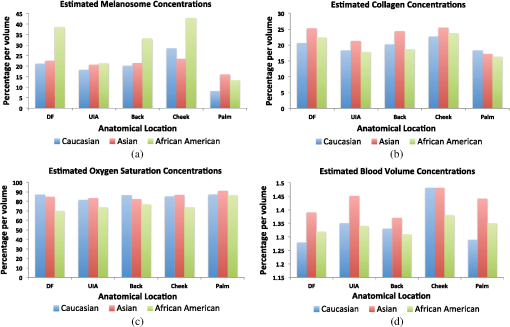 Table 4Root mean square error (RMSE), spectral angle (in radians), and the standard deviation (Std. dev.) for each ethnicity.
Table 5Spectral angle error (in radians) for each anatomical location using the k-NN algorithm.
Fig. 3Plotted, as examples, are 3 of the 241 in vivo signatures along with their estimated signatures using the -nearest neighbors based regression (-NN) algorithm. The estimated parameters for each example are: for the top trace, (17%, 19%, 83%, 0.86%) for the middle trace, and (29%,31%,79%,0.68%) for the bottom trace. 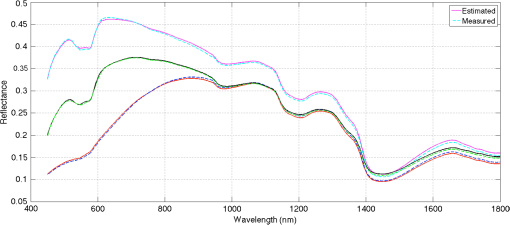 6.DiscussionIn the past 30 years, several studies have attempted to solve the inverse problem of estimating skin parameters from spectral data. The validation of estimated skin parameters is a very difficult task because, in many cases, it is currently impossible to obtain the ground truth. For the case of synthetic skin signatures, the task is simpler, because the ground truth is readily available. It is apparent from Table 2, given the results, that the inverse mapping performs as desired. The task for in vivo signatures is much more difficult because the ground truth is no longer available. Many studies have presented novel inverse methods, but to the best of our knowledge, no studies have augmented their inverse methods with biopsies to ascertain ground truth. A potential validation scheme has often been to check the estimated parameters against well-established physiological precepts. This scheme arose from exploring physiological features that are specific to human skin. For example, as noted by Zonois et al.60 and Nunez,6 melanin is directly responsible for skin color. Therefore, we expect African American subjects to have a larger melanosome concentration than Asian or Caucasian subjects. In a similar manner, there are other physiological precepts that can be checked to ensure that they fit within the realm of physiological plausibility. While this does not guarantee the estimates are accurate, it does offer a good performance validation criteria. Following the precept that skin color correlates with melanosome concentration, it can be seen in the first row of Table 3 and the green bars in Fig. 2 that for each anatomical location, the relative concentration of is the largest for African Americans than for Asians or Caucasians. Furthermore, we would expect higher concentration in the DF and the cheeks, than the palm or the back because these areas are naturally exposed to greater sunlight and hence are inherently more tan. It is clear from Fig. 2 that for caucasian patients, the average concentration is highest in the cheeks at 28.5% and the DF at 21.2%, similarly for Asian and African Americans it is highest in cheeks and the DF at 23.5% and 22.6% and 42.8% and 38.5%, respectively. It is also noteworthy that comparing only cheeks and DF, the relative concentrations are still highest for African Americans followed by Asians and then Caucasian patients. Physiology dictates that we should expect lower concentrations in the palm than the other four areas imaged. It is clear in Fig. 2 that the lowest concentration is in the palm than any other area. There are a number of studies that estimate , and our results are consistent with many of those studies. As such, Zhai et al.61 report concentration of approximately 15% for Caucasians. Our estimate of approximately 18% is in agreement, where the 3% deviation is negligible, considering the variability in skin tone (e.g., tanning, etc.) amongst individuals for each ethnicity. Typically, collagen concentration is higher in the cheeks and the DF than other anatomical locations.61,62 It can be seen in Fig. 2 that the largest collagen concentration for each ethnicity is in the cheeks followed by the DF. There is no pattern for collagen based on ethnicity. We note that the relative collagen concentration between ethnicities remains constant. All patients involved in this study were healthy, so we would expect oxygen saturation concentrations to be greater than 70%. Table 3 shows that the relative oxygen saturation concentrations are above 70%. Kelly et al.,63 Yudovsky and Pilon30 and Tuchin et al.13 have noted that blood volume varies based on anatomical location. It is expected that the blood volume is larger in the cheeks than the DF or the UIA. These precepts are consistent with what is observed in Fig. 2. In summary, the concentrations for all estimated parameters should be physiologically meaningful, i.e., within the acceptable physiological range as outlined in Table 1. This requirement is satisfied as seen by comparing Tables 1 and 3. While these results are encouraging, it is important to note that while all efforts were made to collect hyperspectral data from macroscopically homogeneous skin, a minority of patients had freckles and other types of hyper pigmentation (benign nevi, etc.). This prevented data collection from a completely homogeneous patch of skin. It is possible that these pigmentations contribute some degree of error to our results. Another potential method to validate the estimated parameters is to test the inverse-forward modeling loop itself. In other words, the measured ground truth spectra can be compared to the estimated spectra. While this does not guarantee that the estimated parameters are accurate, it provides some encouraging evidence that the inverse-forward modeling loop works. In this regard, the better the forward model, the higher the probability that the estimated parameters are accurate. Figure 3 shows that the estimated spectra and the ground truth spectra are in good agreement with each other. This performance validation metric is further quantified in Tables 4 and 5, which provide the spectral angles and RMSE for each patient and each anatomical location. While no claims can be made about the exact accuracy of the estimated parameters, the performance validation done through physiological precept analysis and comparisons of the measured and estimated spectra, provides encouraging evidence that the estimated parameters fall within the realm of physiological plausibility. An important consideration in this study is that we fix the skin thickness and the water percentage in the forward model. The thickness of skin can vary based on anatomical location, age, health, etc. In this study, we chose the thickness values tabulated by Meglinski and Matcher15 and Nunez,6 which were obtained based on weighted population averages for these particular anatomical locations. It appears that this choice still leads to reasonable errors on the modeled signature (see Table 4 and Fig. 3) as well as the estimated underlying parameters for our synthetic experiments (see Table 2). However, taking into account varying skin thickness and performing sensitivity analysis are important endeavors which we intend to pursue in future studies. It must be noted that our approach can be extended to estimate the thickness and the water percentage of each of the nine layers of skin. Indeed our inverse methods based on machine learning can handle these extra parameters. While we cannot compare our results directly to other studies because it is not a one-to-one comparison, i.e., we estimate different parameters, on a different dataset, using a different methodology, we can however compare our methodologies and validation metrics. Table 6 provides a summary of this comparison for a few closely related studies. Yudovsky and Pilon30 use a subset of the same physiological precepts as presented here to judge their performance. The estimated blood level (ranging from approximately 1% to 2% by volume in the dermis) is in agreement with this study. While the melanin concentration is not provided as a fraction per volume, if average density for melanin is assumed,64 the estimates are consistent (ranging from approximately 20% to 40% by volume in the epidermis). Cotton24 and Claridge and Preece26,27 perform validation by computing errors associated with their RGB and optimal filters. This is not too different from comparing modeled and estimated signatures, as is done in this study. Effectively, the ground truth is being compared with its estimate in another vector space (rather than in the parameter space). Claridge et al. also employ a modified K-M model with four layers (in contrast to the ten layers used in this study). The physical values of the parameters are not reported, so no further comparisons can be made. Finally, Doi and Tominaga28 also use a K-M based model and a least squares approach to minimize the error between modeled and estimated spectra. Their analysis is similar to the metric used here to compare estimated spectra to the ground truth. They, however, use least squares to find weights for each parameter, rather than reporting their physical values. Therefore, a direct comparison is not possible. In summary, we have shown that the skin parameters from in vivo patients can be noninvasively estimated using our machine learning methods. We have reported acceptable accuracy as per our validation metrics and the metrics used by various other studies, some of which are summarized in Table 6. Table 6Comparison of parameter estimation methodologies and validation metrics.
In analyzing our results, it can be seen in Table 2 that oxygen saturation concentrations exhibit larger errors than the other parameters. A classic problem that has been discussed recently by Nishidate et al.65 has been that of deoxyhemoglobin overestimation in regions with a high concentration of melanosomes. The results in this study seem to echo these findings, especially in Table 3 when comparing the oxygen saturation in African Americans for high melanin regions, and even so in Asian and Caucasian subjects. These trends are consistent even when compared to nonmachine learning studies, such as Yudovsky and Pilon30 or the ones in Table 6. Similarly, this may explain why the errors in Table 2 for oxygen saturation are larger since the synthetic experiments are composed of a wide range of melanosome concentrations. In this regard, we believe our study can serve as a springboard for further investigating this issue. In particular, as a baseline, our synthetic experiments can be redone, where the error in oxygen saturation is plotted as a function of increasing melanosome concentration in the forward model. This is a potential avenue for future work. Finally, an important consideration before such a method can be translated into clinical use is the computational time complexity. This study currently employs a single point spectroscopic system. For this system, our model and algorithm implemented in Matlab R2012b (Natick, Massachusetts) running on an Intel i3-3225 CPU with 8GB DDR3-1600 RAM, takes approximately 30 s to estimate the parameters for each signature. It is important to note that the code is not optimized to take advantage of the parallel architecture of modern CPUs. In the future, we would like to employ a hyperspectral imager. While this would mean that each pixel would require 30 s to analyze, one could implement this system into C++, or some other compiled language so that the computation time is reduced by a factor of 10 to 20 based on the number of threads on the CPU (a current inexpensive CPU has eight threads). 7.ConclusionIn this article, we present a novel application of using hyperspectral signatures and machine learning in concert for estimating the biological parameters of human skin. We find promising results through synthetic and in vivo experiments, which provide encouraging evidence that our methods can potentially be used to noninvasively estimate the skin parameters from hyperspectral signatures. We also report good agreement with the ground truth, and well-established physiological precepts, as well as comparative performance validation with closely related studies. We propose that the temporal evolution of such constitutive parameters, as would be done in a longitudinal study, could help in noninvasive diagnostics and wound-healing applications, amongst others. Our future work will involve validating our methods on datasets containing signatures from individuals with skin abnormalities. We will also investigate means to acquire ground truth for skin parameters in order to help better benchmark our methods. AppendicesAppendixHere we provide a more detailed review of SVR, originally described in Sec. 4.1 We follow the SVR approach and treatment detailed by Smola et al.49,53 We pick up with Eq. (12) from Sec. 4.1, where the goal is to find a small . This is achieved by minimizing the norm, i.e., .53 This problem can then be written as a constrained optimization problem given by: where and are slack variables to account for infeasible constraints on the problem as per the soft margin loss function. is a strictly positive constant, and it accounts for the degree to which errors larger than are tolerated. This is accounted for by optimizing the soft margin loss setting. The specific function, the -insensitive loss function, is given by: In this case (as is common in most cases), the dimensionality of is much higher than the number of observations, therefore, the optimization problem posed in Eq. (18) can be solved with much more ease in its dual formulation. As done in Ref. 49, a dualization method formulated by Fletcher66 using Lagrangian multipliers is implemented. The first task is to construct a Lagrange function from the primal objective function and its corresponding constraints. As per Refs. 67–69, this function has a saddle point with respect to the primal and dual variables at the solution. The Lagrangian function is then given by: where is the Lagrangian and , , , and are the Langrangian multipliers. The Lagrangian multipliers in Eq. (20) have to be greater than or equal to zero as it is a constraint posed by the optimization problem. Furthermore, as a consequence of the saddle point condition, the partial derivative of with respect to each of the primal variables (, , , and ), is zero. This is seen by the following:Equation (21) can then be substituted into Eq. (18), and the new dual optimization problem can hence be written as: In Eq. (22) the dual variables have been eliminated, and the “support vector expansion,” can then be written as:This analysis is typically used for the linear case of SVR; in this work, the nonlinear case needs to be used. However, the analysis and methodology is largely similar. Kernel methods are used in order to account for nonlinearities. Therefore, the linear dot product is converted into a kernel dot product, given by . Then, the process proceeds in the same manner, and from analogy, it arrives at the following result for the support vector expansion: The complexity of the function’s representation by support vectors, therefore, only depends upon the number of support vectors, and not the dimensionality of the input space, . Finally, can be computed using the KKT conditions.54,55 They state that the product between the dual variables and the constraints must go to zero. This can be seen more formally by the following: Based on these constraints, there can never be a set of dual variables that are both nonzero at the same time. Therefore, the following conditions are imposed on : A more formal treatment of choosing an appropriate is detailed in a technical report by Keerthi et al.70 In this manner, each parameter , from Table 1, is independently regressed from the other parameters. Therefore, each is a scalar, where it is one of the five components of being estimated. This methodology is used in this work in order to first compute, , as given by Eq. (13) and ultimately the desired inverse mapping , as detailed in Eq. (9). A more rigorous formulation of SVR is provided in literature.53,56,57 AcknowledgmentsWe would like to thank T. Colella at the Johns Hopkins University, Applied Physics Laboratory, Laurel, Maryland, Drs. L. Garza and S. Kang at the Johns Hopkins University, Department of Dermatology, Baltimore, Maryland, and Dr. R. Chellappa at the University of Maryland, College Park, Maryland, for help with data collection and valuable discussions regarding this study. This project is supported by the Johns Hopkins University, Applied Physics Laboratory, Science and Technology Research and Development Grants, and the Office of Technology Transfer. ReferencesG. Zonioset al.,
“Melanin absorption spectroscopy: new method for noninvasive skin investigation and melanoma detection,”
J. Biomed. Opt., 13
(1), 014017
(2008). http://dx.doi.org/10.1117/1.2844710 JBOPFO 1083-3668 Google Scholar
D. Dickeret al.,
“Differentiation of normal skin and melanoma using high resolution hyperspectral imaging,”
Cancer Biol. Ther., 5
(8), 1033
–1038
(2006). CBTAAO 1538-4047 Google Scholar
M. J. C. van Gemertet al.,
“Skin optics,”
IEEE Trans. Biomed. Eng., 36
(12), 1146
–1154
(1989). http://dx.doi.org/10.1109/10.42108 IEBEAX 0018-9294 Google Scholar
“Cancer facts and figures,”
(2010). Google Scholar
E. M. WurmH. P. Soyer,
“Scanning for melanoma,”
Australian Prescriber, 33
(5), 150
–155
(2010). Google Scholar
A. Nunez,
“A physical model of human skin and its application for search and rescue,”
(2009). Google Scholar
S. Vyaset al.,
“Computational modeling of skin reflectance spectra for biological parameter estimation through machine learning,”
Proc. SPIE, 8390 83901B
(2012). http://dx.doi.org/10.1117/12.919800 PSISDG 0277-786X Google Scholar
S. Vyaset al.,
“Hyperspectral signature analysis of skin parameters,”
Proc. SPIE, 8670 867002
(2013). http://dx.doi.org/10.1117/12.2001428 PSISDG 0277-786X Google Scholar
S. VyasA. BanerjeeP. Burlina,
“Machine learning methods for in vivo skin parameter estimation,”
in Proc. 26th Int. Symp. on Computer-Based Medical Systems,
(2013). Google Scholar
G. BaranoskiA. Krishnaswamy,
“Light interaction with human skin: from believable images to predictable models,”
in Proc. ACM SIGGRAPH ASIA’08 courses,
(2008). Google Scholar
P. KubelkaF. Munk,
“An article on optics of paint layers,”
Zeitschrift für Technische Physik, 12 593
–601
(1931). ZTPHAU 0373-0093 Google Scholar
M. van GemertW. Star,
“Relations between the Kubelka-Munk and the transport equation models for anisotropic scattering,”
Lasers Life Sci., 1
(98), 287
–298
(1987). Google Scholar
V. TuchinS. UtzI. Yaroslavsky,
“Tissue optics, light distribution, and spectroscopy,”
Opt. Eng., 33
(10), 3178
–3188
(1994). http://dx.doi.org/10.1117/12.178900 OPEGAR 0091-3286 Google Scholar
G. Yoon,
“Absorption and scattering of laser light in biological media-mathematical modeling and methods for determining the optical properties,”
(1988). Google Scholar
I. V. MeglinskiS. J. Matcher,
“Quantitative assessment of skin layers absorption and skin reflectance spectra simulation in the visible and near-infrared spectral regions,”
Physiol. Meas., 23
(4), 741
–753
(2002). http://dx.doi.org/10.1088/0967-3334/23/4/312 PMEAE3 0967-3334 Google Scholar
A. Ishimaru, Wave Propagation and Scattering in Random Media, Wiley-IEEE Press, New York
(1999). Google Scholar
S. Prahl,
“Light transport in tissue,”
University of Texas,
(1988). Google Scholar
H. Van de Hulst,
“Multiple light scattering: tables,”
Formulas and Applications, 477
–492 Academic Press, New York
(1980). Google Scholar
D. Churmakovet al.,
“Analysis of skin tissues spatial fluorescence distribution by the Monte Carlo simulation,”
J. Phys. D Appl. Phys., 36
(14), 1722
–1728
(2003). http://dx.doi.org/10.1088/0022-3727/36/14/311 JPAPBE 0022-3727 Google Scholar
M. Shimadaet al.,
“Melanin and blood concentration in a human skin model studied by multiple regression analysis: assessment by Monte Carlo simulation,”
Phys. Med. Biol., 46
(9), 2397
–2406
(2001). http://dx.doi.org/10.1088/0031-9155/46/9/309 PHMBA7 0031-9155 Google Scholar
P. HanrahanW. Krueger,
“Reflection from layered surfaces due to subsurface scattering,”
in Proc. 20th Annual Conf. on Computer Graphics and Interactive Techniques,
165
–174
(1993). Google Scholar
C. So-LingL. Li,
“A multi-layered reflection model of natural human skin,”
in Proc. Computer Graphics Int.,
249
–256
(2001). Google Scholar
J. Stam,
“An illumination model for a skin layer bounded by rough surfaces,”
in Proc. 12th Eurographics Workshop on Rendering Techniques,
39
–52
(2001). Google Scholar
S. Cotton,
“A noninvasive skin imaging system,”
(1997). Google Scholar
S. CottonE. Claridge,
“Developing a predictive model of human skin colouring,”
Proc. SPIE, 2708 814
–825
(1996). http://dx.doi.org/10.1117/12.237846 PSISDG 0277-786X Google Scholar
E. ClaridgeS. Preece,
“An inverse method for the recovery of tissue parameters from colour images,”
Information Processing in Medical Imaging, 306
–317 Springer, New York
(2003). Google Scholar
S. J. PreeceE. Claridge,
“Spectral filter optimization for the recovery of parameters which describe human skin,”
IEEE Trans. Pattern Anal. Mach. Intell., 26
(7), 913
–922
(2004). http://dx.doi.org/10.1109/TPAMI.2004.36 ITPIDJ 0162-8828 Google Scholar
M. DoiS. Tominaga,
“Spectral estimation of human skin color using the Kubelka-Munk theory,”
Proc. SPIE, 5008 221
–228
(2003). http://dx.doi.org/10.1117/12.472026 PSISDG 0277-786X Google Scholar
D. YudovskyL. Pilon,
“Rapid and accurate estimation of blood saturation, melanin content, and epidermis thickness from spectral diffuse reflectance,”
Appl. Opt., 49
(10), 1707
–1719
(2010). http://dx.doi.org/10.1364/AO.49.001707 APOPAI 0003-6935 Google Scholar
D. YudovskyL. Pilon,
“Retrieving skin properties from in vivo spectral reflectance measurements,”
J. Biophoton., 4
(5), 305
–314
(2011). http://dx.doi.org/10.1002/jbio.201000069 JBOIBX 1864-063X Google Scholar
R. AndersonJ. Parrish,
“The optics of human skin,”
J. Invest. Dermatol., 77
(1), 13
–19
(1981). http://dx.doi.org/10.1111/jid.1981.77.issue-1 JIDEAE 0022-202X Google Scholar
S. WanR. AndersonJ. Parrish,
“Analytical modeling for the optical properties of the skin with in vitro and in vivo applications,”
Photochem. Photobiol., 34
(4), 493
–499
(1981). http://dx.doi.org/10.1111/j.1751-1097.1981.tb09030.x PHCBAP 0031-8655 Google Scholar
N. TsumuraH. HaneishiY. Miyake,
“Independent-component analysis of skin color image,”
J. Opt. Soc. Am. A, 16
(9), 2169
–2176
(1999). http://dx.doi.org/10.1364/JOSAA.16.002169 JOAOD6 0740-3232 Google Scholar
N. Tsumuraet al.,
“Image-based skin color and texture analysis/synthesis by extracting hemoglobin and melanin information in the skin,”
ACM Trans. Graph., 22
(3), 770
–779
(2003). http://dx.doi.org/10.1145/882262.882344 ATGRDF 0730-0301 Google Scholar
N. Tsumuraet al.,
“Mapping pigmentation in human skin from a multi-channel visible spectrum image by inverse optical scattering technique,”
J. Imag. Sci. Technol., 45
(5), 444
–450
(2001). JIMTE6 1062-3701 Google Scholar
E. Claridgeet al.,
“From colour to tissue histology: physics-based interpretation of images of pigmented skin lesions,”
Med. Image Anal., 7
(4), 489
–502
(2003). http://dx.doi.org/10.1016/S1361-8415(03)00033-1 MIAECY 1361-8415 Google Scholar
I. Kaartinenet al.,
“How to assess scar hypertrophya—a comparison of subjective scales and Spectrocutometry: a new objective method,”
Wound Repair Regen., 19
(3), 316
–323
(2011). http://dx.doi.org/10.1111/j.1524-475X.2011.00679.x WREREU 1067-1927 Google Scholar
L. Leonardiet al.,
“Near-infrared spectroscopy and imaging: a new approach to assess burn injuries,”
Am. Clin. Lab., 19
(8), 20
–22
(2000). ACLAE7 1041-3235 Google Scholar
J. Payetteet al.,
“Assessment of skin flaps using optically based methods for measuring blood flow and oxygenation,”
Plastic Reconstruc. Surg., 115
(2), 539
–546
(2005). http://dx.doi.org/10.1097/01.PRS.0000148415.54546.CA PRSUAS 0032-1052 Google Scholar
S. Prigentet al.,
“Multi-spectral image analysis for skin pigmentation classification,”
in Proc. 17th IEEE Int. Conf. on Image Process.,
3641
–3644
(2010). Google Scholar
S. Prigentet al.,
“Spectral analysis and unsupervised SVM classification for skin hyper-pigmentation classification,”
in Proc. 2nd Workshop on Hyperspectral Image and Signal Process.: Evolution in Remote Sensing,
1
–4
(2010). Google Scholar
D. YudovskyA. NouvongL. Pilon,
“Hyperspectral imaging in diabetic foot wound care,”
J. Diabetes Sci. Technol., 4
(5), 1099
–1113
(2010). Google Scholar
S. Dreiseitlet al.,
“A comparison of machine learning methods for the diagnosis of pigmented skin lesions,”
J. Biomed. Inform., 34
(1), 28
–36
(2001). http://dx.doi.org/10.1006/jbin.2001.1004 JBIOBL 1532-0464 Google Scholar
J. Nyströmet al.,
“Combined near-infrared spectroscopy and multifrequency bio-impedance investigation of skin alterations in diabetes patients based on multivariate analyses,”
Med. Biol. Eng. Comput., 41
(3), 324
–329
(2003). http://dx.doi.org/10.1007/BF02348438 MBECDY 0140-0118 Google Scholar
M. Sowaet al.,
“Near infrared spectroscopic assessment of hemodynamic changes in the early post-burn period,”
Burns, 27
(3), 241
–249
(2001). http://dx.doi.org/10.1016/S0305-4179(00)00111-X BURND8 0305-4179 Google Scholar
V. Bochkoet al.,
“Lower extremity ulcer image segmentation of visual and near-infrared imagery,”
Skin Res. Technol., 16
(2), 190
–197
(2010). http://dx.doi.org/10.1111/srt.2010.16.issue-2 0909-752X Google Scholar
P. GeladiD. MacDougallH. Martens,
“Linearization and scatter-correction for near-infrared reflectance spectra of meat,”
Appl. Spectrosc., 39
(3), 491
–500
(1985). http://dx.doi.org/10.1366/0003702854248656 APSPA4 0003-7028 Google Scholar
I. MeglinskiS. Matcher,
“Modeling of skin reflectance spectra,”
Proc. SPIE, 4241 78
–87
(2001). http://dx.doi.org/10.1117/12.431508 PSISDG 0277-786X Google Scholar
V. VapnikS. GolowichA. Smola,
“Support vector method for function approximation, regression estimation and signal processing,”
in Proc. Adv. in Neural Information Process. Syst. Conf.,
281
–287
(1996). Google Scholar
C. CortesV. Vapnik,
“Support-vector networks,”
Mach. Learn., 20
(3), 273
–297
(1995). http://dx.doi.org/10.1023/A:1022627411411 MALEEZ 0885-6125 Google Scholar
A. Navotet al.,
“Nearest neighbor based feature selection for regression and its application to neural activity,”
in Proc. Adv. in Neural Information Process. Syst. Conf.,
995
–1002
(2006). Google Scholar
C.-C. ChangC.-J. Lin,
“LIBSVM: a library for support vector machines,”
ACM Trans. Intell. Syst. Technol., 2
(3), 27
(2011). http://dx.doi.org/10.1145/1961189.1961199 Google Scholar
A. SmolaB. Scholkopf,
“A tutorial on support vector regression,”
Stat. Comput., 14
(3), 199
–222
(2004). http://dx.doi.org/10.1023/B:STCO.0000035301.49549.88 STACE3 0960-3174 Google Scholar
W. Karush,
“Minima of functions of several variables with inequalities as side constraints,”
University of Chicago,
(1939). Google Scholar
H. KuhnA. Tucker,
“Nonlinear programming,”
in Proc. 2nd Berkeley Symposium on Mathematical Statistics and Probability,
481
–492
(1951). Google Scholar
R. CollobertS. Bengio,
“SVMTorch: support vector machines for large-scale regression problems,”
J. Mach. Learn. Res., 1 143
–160
(2001). http://dx.doi.org/10.1162/15324430152733142 1532-4435 Google Scholar
C. Burges,
“A tutorial on support vector machines for pattern recognition,”
Data Min. Knowl. Discov., 2
(2), 121
–167
(1998). http://dx.doi.org/10.1023/A:1009715923555 1384-5810 Google Scholar
G. Bradski,
“The OpenCV Library,”
Dr. Dobb’s J. Softw. Tools, 25
(11), 120
–126
(2000). Google Scholar
M. Tommolaet al.,
“Estimating the characteristics of a marked stand using k-nearest-neighbour regression,”
Int. J. Forest Eng., 10
(2), 75
–81
(1999). Google Scholar
G. ZoniosJ. BykowskiN. Kollias,
“Skin melanin, hemoglobin, and light scattering properties can be quantitatively assessed in vivo using diffuse reflectance spectroscopy,”
J. Invest. Dermatol., 117
(6), 1452
–1457
(2001). http://dx.doi.org/10.1046/j.0022-202x.2001.01577.x JIDEAE 0022-202X Google Scholar
H. ZhaiK. WilhelmH. Maibach, Dermatotoxicology, Informa Healthcare(2007). Google Scholar
C. Lovellet al.,
“Type I and III collagen content and fibre distribution in normal human skin during ageing,”
Br. J. Dermatol., 117
(4), 419
–428
(1987). http://dx.doi.org/10.1111/j.1365-2133.1987.tb04921.x BJDEAZ 1365-2133 Google Scholar
R. Kellyet al.,
“The effects of aging on the cutaneous microvasculature,”
J. Am. Acad. Dermatol., 33
(5), 749
–756
(1995). http://dx.doi.org/10.1016/0190-9622(95)91812-4 JAADDB 0190-9622 Google Scholar
T. Dwyeret al.,
“Melanin density and melanin type predict melanocytic naevi in 19-20 year olds of northern European ancestry,”
Melanoma Res., 10
(4), 387
–394
(2000). MREEEH 0960-8931 Google Scholar
I. Nishidateet al.,
“Noninvasive imaging of human skin hemodynamics using a digital red-green-blue camera,”
J. Biomed. Opt., 16
(8), 086012
(2011). http://dx.doi.org/10.1117/1.3613929 JBOPFO 1083-3668 Google Scholar
R. Fletcher, Practical Methods of Optimization, Wiley-Interscience, New York
(1987). Google Scholar
O. Mangasarian, Nonlinear Programming, Society for Industrial Mathematics, New York
(1994). Google Scholar
G. McCormick, Nonlinear Programming: Theory, Algorithms, and Applications, John Wiley & Sons, Inc., New York
(1983). Google Scholar
R. J. VanderbeiR. J. Vanderbei,
“LOQO users manualversion 3.10,”
Optimiz. Methods Softw., 11 485
–514
(1999). Google Scholar
S. Keerthiet al.,
“Improvements to Platt’s SMO algorithm for SVM classifier design,”
Neural Comput., 13
(3), 637
–649
(2001). http://dx.doi.org/10.1162/089976601300014493 NEUCEB 0899-7667 Google Scholar
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

