|
|
|
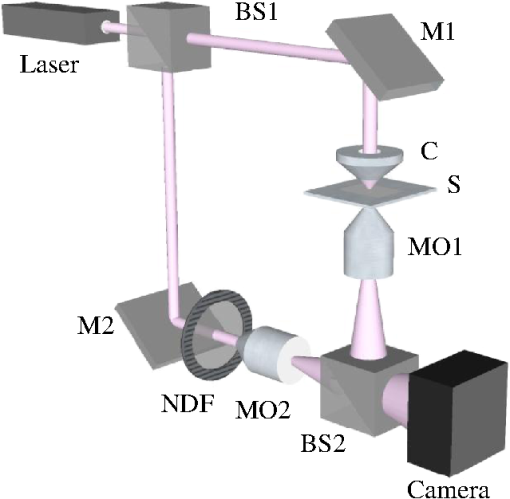
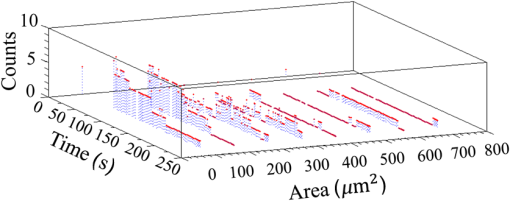
Epidemiology of several important human diseases, such as atherosclerosis, Alzheimer’s disease, and cancer, contains a lipid component.1–3 Moreover, it is known that the lipid composition of membranes and membrane–protein adsorption mechanisms are connected, and this connection affects the intermembrane spacing and adhesion and has direct effect on demyelinating diseases such as multiple sclerosis.4 In general, information on the structure of membranes including the dynamic organization across the lipid bilayer and in the lateral dimension to create unique composition of lipids with multiple functionalities have been of particular importance for biochemical and biophysical investigations.5–7 Smectic liquid-crystalline (LC) order is spontaneously created in multilamellae stacked lipids, as lyotropic materials, through hydrophobic forces of lipid–water interactions.8–10 Moreover, these materials show long-ranged orientation order in the presence of a solvent in LC phases.11 In smectic mesophases materials, including multilamellae stacked lipids, in-plane translational degrees of freedom are allowed, but their out-of-plane undulatory fluctuations are impeded due to their confinement by the near-located layers.11–13 One-dimensional smectic order lipidic mesophases can be stabilized by losing entropy through Helfrich intermembrane repulsive interactions.14 Studies on ordering mechanisms of such membrane multilamellae find useful applications in the field of membrane-based functional materials. Functional membranes offer innumerable possible applications such as pharmaceutical purification processes,15,16 organic solvent filtration,17 sustainable energy production,18 and high-performance filtration under harsh conditions.19,20 Functional multilamellae membranes can be resembled by a multicomponent mixture of lipids and proteins, which contains coexisting phases from facile phase separations.11 In these membranes, two-dimensional (2-D) intralayer phase separation of lipids and proteins in individual membrane lamellae can be combined with the interlayer smectic ordering, which leads to arranged functional components across membrane lamellae in a self-organized manner.11,21 Higher order organization in multilayered membranes has been observed by conventional wide-field epifluorescence microscopy.11 In this paper, a quantitative study on volumetric dynamics of multilamellar multicomponent membranes is considered. We used digital holographic microscopy (DHMicroscopy) to evaluate thickness, surface area, and volume of the domains extracted from phase separation in multicomponent lipids versus time. DHMicroscopy is a nondestructive and marker-free imaging technique suitable for three-dimensional (3-D) visualization of phase objects.22–27 The interference pattern of a coherent beam passing through the object with a reference beam from the same source forms a hologram on a digital camera. The reconstruction process toward acquiring a 3-D image of the object is performed by numerical reconstruction of the digital holograms by a computer.28–31 The samples we used for membrane multilamellae are nonuniform systems consisting of more than a single lipid. They are lipid mixtures composed of cholesterol (Chol), sphingomyelin (SM), and unsaturated phospholipid, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), as putative raft-forming ternary. This kind of rafts or domains in biomembranes is used for signaling activities. For example, ion channels are localized in liquid-ordered (Lo) domains and the alignment of domains also aligns the ion channels. The alignment of domains across several bilayers may be used for coupling of the functionality of single bilayers. We purchased the lipids in powder type from Avanti, and chloroform was used as the solvent. Clean and dry glass slides were used during the experiments. Droplets with the volume of 1 to of the solution were put on a glass plate. The samples were kept in a vacuum overnight for chloroform evaporation and then were incubated at 50°C to 60°C in a humid environment with relative humidity of 98% for 48 to 72 h. For each experiment, by fast immersion of dried plaques of multilamellar lipids in deionized bulk water, the phase separating regime was achieved from quenching the high temperature of the homogeneous phase. The whole process was observed using a DHMicroscopy setup in which a laser beam illuminates the object and the magnification is provided by the microscope objective lens and imaging lens combination. The DHMicroscopy setup is based on Mach–Zehnder geometry to allow off-axis holography of a transparent sample, as shown schematically in Fig. 1. On the same setup, a conventional homemade inverted microscope was built; it consists of a white light illumination source (KL1500 compact, Olympus), condenser (C, NA 1.25, Olympus), microscope objective (MO1, , NA 0.65, WD 0.17 mm, Olympus), and the camera (DCC1545M, Thorlabs, 8-bit dynamic range, pixel pitch). The samples were observed through the microscope to adjust the best focus before the holography experiments. Laser light emitted from a He–Ne laser (MEOS, 632.8 nm, 5 mW) is expanded by a beam expander and is split into object wave and reference wave by a beam splitter (BS1). The reference beam is sent to the camera through mirror M1, a similar microscope objective (MO2), which was used to adjust the beam curvature, and a beam splitter (BS2). The reference beam is focused at a point conjugate to the back focal plane of the objective lens so that it arrives at the detector plane with the same wave front curvature as the object wave. Digital holograms that are formed by the interference pattern of the two beams have 3-D information of the sample under study. The numerical reconstruction process is performed by simulating the diffraction from the digital hologram when illuminated by the reference wave. Previously, we have utilized similar DHMicroscopy setups and also self-referencing DHMicroscopy scheme for 3-D imaging and investigating lipids.32–34 In Ref. 32, the effect of thermal gradient on multilamellar cylindrical tubes of membrane lipids, which are called myelin figures, has been investigated. In Ref. 33, we have shown the lipid resource humidifying impact on the growth control of myelin figures. In Ref. 34, we have used a common-path self-referencing DHMicroscopy arrangement coupled to a fiber optical trapping system to induce morphological changes on lipid membranes by the forces applied by an optical fiber with an enhanced numerical aperture. In our experiments, we recorded the holograms of the samples of multicomponent lipids by a Mach–Zehnder-based DHMicroscopy setup. The holograms were recorded in video format and were converted into image sequences for postprocessing. The lipid mixtures are phase-separated into two coexisting liquid phases below their miscibility transition temperature. One of the phases is enriched in Chol and SM, designated as the Lo phase, and the other is the liquid-disordered (Ld) phase consisting primarily of unsaturated DOPC.11,35 Since the incubation of the lipids were performed in the range of 50°C and 60°C, and this range is above the miscibility transition temperatures of the lipid mixtures, the separated domains are not formed during the incubation. Phase-separated domains in individual layers of multilamellar membrane stacks align with one another, spontaneously producing 3-D aligned domains across hundreds of lamellae.11 Figure 2 shows a selected set of frames from conventional microscopy imaging of examination of the domain formation and domain surface size evolution, immediately after immersing the multilamellar lipids cakes in bulk water. The lipid mixture consists of equimolar proportions of DOPC and SM and 30% of Chol. We have also shown that in higher Chol concentrations the domain alignment process is substantially reduced, and in more than 40% concentrations, the domains turn to appear as crystals, as shown in Fig. 3. Fig. 2Bright-field images of phase separation in stacked lipid mixture, consisting of equimolar proportions of DOPC and SM, and 30% of Chol immersed in bulk water. Domain evolution is shown for various times after immersion. 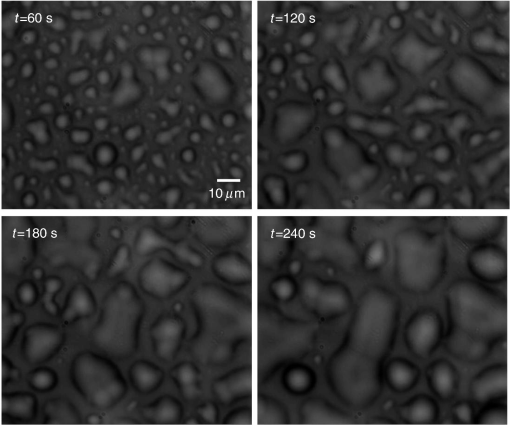 Fig. 3Up to 40% of Chol concentrations coexisting Lo and Ld phases were observed. However, in composition with 50% and 60% concentrations of Chol, materials appear as crystals, as shown in these figures. 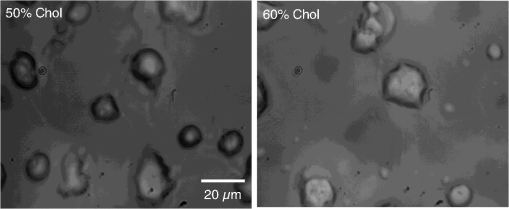 However, toward a better examination of interlamellar domain alignment and phase separation in multicomponent lipids, we utilized DHMicroscopy and investigated the thickness profile of samples. This, in turn, provides quantitative volumetric information on the evolution of the formed domains and alignment process. In Fig. 4, a digital hologram of a lipid mixture sample when the domains are formed and the associated reference holograms are shown. Smaller regions of interest for both holograms are shown in the lower panel of Fig. 4; it demonstrates the holographic interference pattern formation and fringe inclination changes. We record reference holograms in which no lipid sample is presented to remove the background contaminations from the sample container and the fluid for every sample. This ensures that the phase changes in various times in the domain evolution process are only due to its growth. The recorded holograms are subjected to numerical reconstruction. The reconstruction process is performed by simulating the diffraction from the digital hologram when illuminated by the reference wave. We utilized the angular spectrum propagation approach in scalar diffraction theory for numerical reconstruction of the holograms.28–31 The phase and intensity of the reconstructed wavefront can be computed by the ratio of imaginary and real parts, and the squared absolute value of the derived complex amplitude, respectively. The objects under study in our case are almost transparent; hence, we are interested in measurement of phase changes during the experiments. According to the dynamics speed of the phenomenon, we recorded holographic movies with a rate of in various states of domain formation and evolution. Fig. 4Hologram of a lipid mixture sample when the domains are formed and associated reference holograms. A smaller region of interest for both holograms is shown in the lower panel to demonstrate the holographic intehreference patten formation and fringe inclination changes. 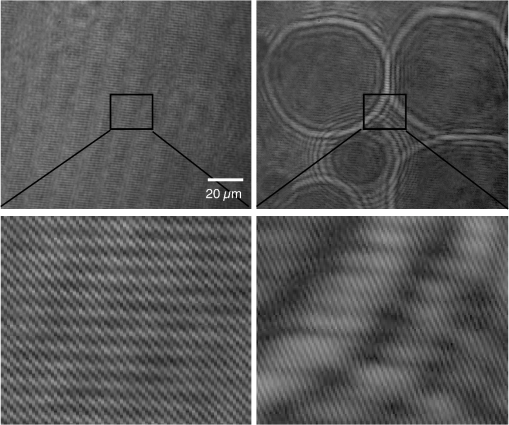 Figure 5 shows the reconstruction results for typical domain evolution that we have observed in multicomponent lipid samples. The holograms are in left and right panels belong to and after immersing the lipid cake in bulk water, respectively. The two formed domains at join together to form a bigger one at . In Figs. 5(a)–5(c), different illustrations of the phase information from the DHMicroscopy reconstruction are shown. Assuming negligible changes for the refractive index, the thickness at each point can be calculated from the phase map. A 3-D map of thickness of the domains in the two chosen times is shown in Fig. 5(a). A vertical view (2-D map) of the two reconstructed images is shown in Fig. 5(b). As can be seen, the phase of the domain and the rest of the field of view have different value ranges. By a simple Red-Green-Blue to binary transformation of the map, the surface covered by any domain can be calculated. The cross sectional thickness profile along any arbitrary line (like ) across the lipid can also be calculated, as shown in Fig. 5(c). Fig. 5Reconstructed holographic images of domain formation in multicomponent lipid samples at and 45 s after the formation starts: (a) 3-D thickness of the domains, (b) 2-D thickness map, and (c) cross sectional profile along the () line indicated on the 2-D images in panel (b). Information such as volume, surface, and thickness of the domains can be derived from the reconstruction of holograms. A field of view of about from the recoreded holograms was selected for measurements. 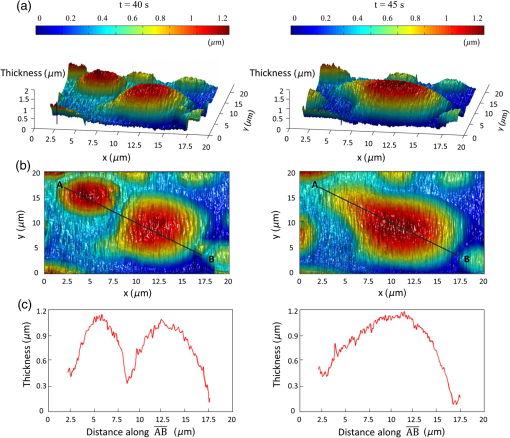 For each frame of the recorded movies, we have reconstructed the holograms, counted the number of formed domains, and calculated the surface, thickness, and volume of the domains. The average volume, surface, and thickness are calculated by averaging the values among all the domains in the field of view of each frame. The thickness is the mean value of the averaged domain thickness. These values are reported and shown in Fig. 6. Figures 6(a)–6(c) are the evolution of the volume, surface, and thickness of domains, respectively, for 4 min after the experiment is started. In the inset of Fig. 6(b), a similar evolution curve for the covered surface by domains, which is obtained from conventional microscopy image analysis, is shown. Fig. 6Evolution of domain size in stacked multicomponent lipid bilayers including of equimolar SM and DOPC1 and 30% Chol: (a) relative volume of domains extracted from averaging several domains in each acquired image, calculated through digital holograms reconstruction; (b) evolution of average area covered by domains versus time, obtained from DHMicroscopy. Inset: Similar curve obtained from conventional microscopy images analysis; and (c) evolution of relative thickness averaged for several domains in each time. The universal growth law as have been fitted in all cases, showing the similar growth behavior for volume and area, different from thickness behavior. 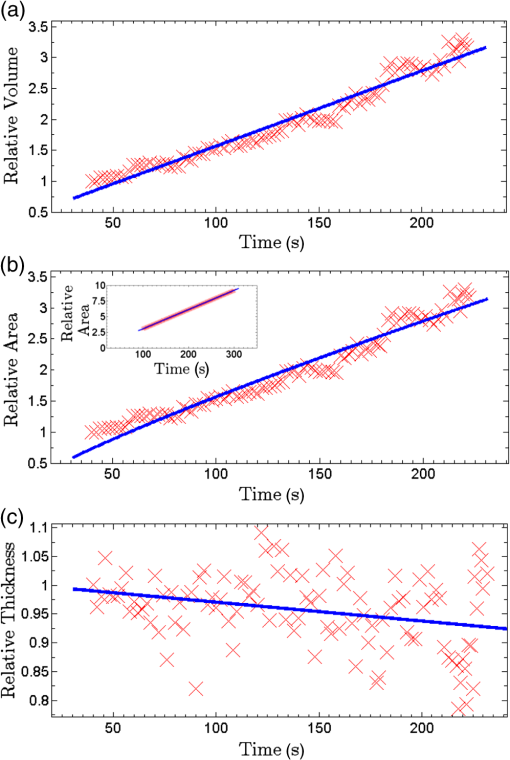 The temporal evolution of circular domain size (average area) in single lipid monolayers or bilayers follows the universal growth law as .36 According to classical diffusion, the limited growth . It has been proven that in membrane multilamellae the domain growth also follows the universal growth law.11 However, in this case, the growth exponent varies between and . The higher exponents suggest that the domain growth involves additional contributions beyond diffusional dynamics, such as the domain–domain coalescence as shown for a typical sample in Fig. 5. The coalescence-driven domain growth results from an interplay between lateral phase separation and interlamellar smectic coupling unique to membrane multilamellae.11 Accordingly, we have fitted the universal growth law for the all extracted sizes of domains including volume, area, and thickness. As shown in Fig. 6, the growth of domain volume and area follows similar behavior. For a typical sample after averaging all the analyzed domains of the holograms, we found that volume and area varies as and , respectively. The growth component for area evolution is iconsistent with the results of bright field microscopy analysis, as shown in the inset of Fig. 6(b). However, we have observed that even if the volume of domains follows the universal growth law, as shown in Fig. 6(c), the change in volume should be attributed mostly to the changes in area of the domains, and the average thickness of the domains does not have a substantial variation versus time. Yet, for several cases, the thickness can be reduced even at longer times after the experiment is started, which explains the difference between volume and area exponents. The evolution of the domain sizes was calculated and sketched for the average of the domain sizes within the field of view of each frame of the movies acquired through the DHMicroscopy setup. Further, we have shown the histogram of the domain area size versus time in Fig. 7. As the histograms show, in the initial seconds of the experiments, the number of domains with a smaller size is greater than the larger ones. However, while time passes, the number of large sized domains increases and, in contrast, the number of small ones decreases. This proves the idea of the effective role of the domain–domain coalescence on the domain size growth. In conclusion, we have studied the coupling between intralayer phase separations and interlayer LC ordering in multicomponent membranes. We have used DHMicroscopy for volumetric measurement of the phenomenon dynamics. The samples include mixed species of lipids and cholesterol. DHMicroscopy provides detailed information on the thickness profile and volumes of the lipids. Our results show that the volume evolution of lipid domains follows approximately the same universal growth law of area evolution. However, their thickness will not change significantly by time. The volume evolution is mostly attributed to the changes in area dynamics. We believe that our results can be useful in the field of membrane-based functional materials, and the approach of DHMicroscopy has the potential to be utilized for similar problems in the field of membranes. DisclosuresThe authors have no relevant financial interests and no other potential conflicts of interest. ReferencesF. R. Maxfield and I. Tabas,
“Role of cholesterol and lipid organization in disease,”
Nature, 438 612
–621
(2005). http://dx.doi.org/10.1038/nature04399 Google Scholar
A. Helgadottir et al.,
“Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease,”
Nat. Genet., 48 634
–639
(2016). http://dx.doi.org/10.1038/ng.3561 Google Scholar
N. E. Braverman and A. B. Moser,
“Functions of plasmalogen lipids in health and disease,”
Biochim. Biophys. Acta, 1822 1442
–1452
(2012). BBALAJ 0067-2734 Google Scholar
D. W. Lee et al.,
“Lipid domains control myelin basic protein adsorption and membrane interactions between model myelin lipid bilayers,”
Proc. Natl. Acad. Sci. U. S. A., 111 E768
–E775
(2014). http://dx.doi.org/10.1073/pnas.1401165111 Google Scholar
G. van Meer, D. R. Voelker and G. W. Feigenson,
“Membrane lipids: where they are and how they behave,”
Nat. Rev. Mol. Cell Biol., 9 112
–124
(2008). http://dx.doi.org/10.1038/nrm2330 NRMCBP 1471-0072 Google Scholar
C. Eggeling et al.,
“Direct observation of the nanoscale dynamics of membrane lipids in a living cell,”
Nature, 457 1159
–1162
(2008). http://dx.doi.org/10.1038/nature07596 Google Scholar
L. Chernomordik, M. M. Kozlov and J. Zimmerberg,
“Lipids in biological membrane fusion,”
J. Membr. Biol., 146 1
–14
(1995). http://dx.doi.org/10.1007/BF00232676 Google Scholar
R. G. Laughlin, The Aqueous Phase Behaviour of Surfactants, Academic Press, London
(1996). Google Scholar
D. F. Evans and H. Wennerstrom, The Colloidal Domain: Where Physics, Chemistry, Biology and Technology Meet, Wiley, New York
(1999). Google Scholar
Y. K. Levine, A. B. Bailey and M. H. F. Wilkins,
“Multilayers of phospholipid bimolecular leaflets,”
Nature, 220 577
–578
(1968). http://dx.doi.org/10.1038/220577a0 Google Scholar
L. Tayebi et al.,
“Long-range interlayer alignment of interlayer domains in stacked lipid bilayer,”
Nat. Mater., 11
(12), 1074
–1080
(2012). http://dx.doi.org/10.1038/nmat3451 Google Scholar
J. Yamamoto and H. Tanaka,
“Dynamic control of the photonic smectic order of membranes,”
Nat. Mater., 4 75
–80
(2005). http://dx.doi.org/10.1038/nmat1281 Google Scholar
P. F. F. Almeida,
“Thermodynamics of lipid interactions in complex bilayers,”
Biochim. Biophys. Acta, 1788
(1), 72
–85
(2009). http://dx.doi.org/10.1016/j.bbamem.2008.08.007 BBACAQ 0006-3002 Google Scholar
P. Sharma,
“Entropic force between membranes reexamined,”
Proc. Natl. Acad. Sci. U. S. A., 110
(6), 1976
–1977
(2013). http://dx.doi.org/10.1073/pnas.1222033110 Google Scholar
K. V. Peinmann and S. P. Nunens, Membranes for the Life Sciences, Wiley, New York
(2008). Google Scholar
L. Tayebi, D. Vashaee and A. N. Parikh,
“Stability of uni- and multillamellar spherical vesicles,”
Chem. Phys. Chem., 13
(1), 314
–322
(2012). http://dx.doi.org/10.1002/cphc.201100573 Google Scholar
K. M. Sivaraman et al.,
“Porous polysulfone coating for enhanced drug delivery,”
Biomed. Microdevice, 14 603
–612
(2012). http://dx.doi.org/10.1007/s10544-012-9639-6 BMICFC 1387-2176 Google Scholar
P. Marchetti et al.,
“Molecular separation with organic solvent nonfilteration: a critical review,”
Chem. Rev., 114
(21), 10735
–10806
(2014). http://dx.doi.org/10.1021/cr500006j CHREAY 0009-2665 Google Scholar
J. A. Kinler et al., Functional Material for Sustainable Energy Applications, Woodhead Publishing, Cambridge, United Kingdom
(2012). Google Scholar
Y. Zhu et al.,
“Recent progress in developing advanced membranes for emulsified oil/water separation,”
NPG Asia Mater., 6 e101
(2014). http://dx.doi.org/10.1038/am.2014.23 1884-4057 Google Scholar
Y. Zhu, A. Negmi and J. Moran-Mirabel,
“Multi-stacked supported lipid bilayer micropatterning through polymer stencil lift-off,”
Membranes, 5
(3), 385
–398
(2015). http://dx.doi.org/10.3390/membranes5030385 MBRNAR 0076-6356 Google Scholar
U. Schanars and W. Japtner,
“Digital recording and numerical reconstruction of holograms,”
Meas. Sci. Tech., 13 R85
–R101
(2002). http://dx.doi.org/10.1088/0957-0233/13/9/201 MSTCEP 0957-0233 Google Scholar
M. K. Kim,
“Principle and techniques of digital holographic microscopy,”
SPIE Rev., 1
(1), 018005
(2010). http://dx.doi.org/10.1117/6.0000006 Google Scholar
I. Moon et al.,
“Cell identification computational 3-D holographic microscopy,”
Opt. Photonics News, 22
(6), 18
–23
(2011). http://dx.doi.org/10.1364/OPN.22.6.000018 OPPHEL 1047-6938 Google Scholar
P. Marquet et al.,
“Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with sub-wavelength axial accuracy,”
Opt. Lett., 30 468
–470
(2005). http://dx.doi.org/10.1364/OL.30.000468 OPLEDP 0146-9592 Google Scholar
Y. Frauel et al.,
“Three-dimensional imaging and processing using computational holographic imaging,”
Proc. IEEE, 94
(3), 636
–653
(2006). http://dx.doi.org/10.1109/JPROC.2006.870704 Google Scholar
V. Bianco et al.,
“Clear microfluidics imaging through flowing blood by digital holography,”
IEEE J. Sel. Top. Quantum Electron., 20
(3), 6801507
(2014). http://dx.doi.org/10.1109/JSTQE.2013.2286075 Google Scholar
D. Shin et al.,
“Optofluidic system for three-dimensional sensing and identification of micro-organisms with digital holographic microscopy,”
Opt. Lett., 35 4066
–4068
(2010). http://dx.doi.org/10.1364/OL.35.004066 OPLEDP 0146-9592 Google Scholar
W. Goodman, Introduction to Fourier Optics, McGraw-Hill, New York
(2005). Google Scholar
F. Zhang, G. Pedrini and W. Osten,
“Reconstruction algorithm for high-numerical-aperture hologram with diffraction-limited resolution,”
Opt. Lett., 31 1633
–1635
(2006). http://dx.doi.org/10.1364/OL.31.001633 OPLEDP 0146-9592 Google Scholar
A. Anand, V. K. Chhaniwal and B. Javidi,
“Real time digital holographic microscopy for phase contras 3D-imaging of dynamic phenomena,”
J. Disp. Tech., 6 500
–505
(2010). http://dx.doi.org/10.1109/JDT.2010.2052020 Google Scholar
N. Fathi et al.,
“Digital holographic microscopy of the myelin figure structural dynamics and the effect of thermal gradient,”
Biomed. Opt. Express, 4 950
–957
(2013). http://dx.doi.org/10.1364/BOE.4.000950 BOEICL 2156-7085 Google Scholar
R. Mosaviani, A. R. Moradi and L. Tayebi,
“Effect of humidity on liquid-crystalline myelin figure growth using digital holographic microscopy,”
Mater. Lett., 173 162
–166
(2016). http://dx.doi.org/10.1016/j.matlet.2016.03.023 Google Scholar
S. Ebrahimi et al.,
“Digital holographic microscopy with coupled optical fiber trap for cell measurement and manipulation,”
Opt. Lett., 39 2916
–2919
(2014). http://dx.doi.org/10.1364/OL.39.002916 OPLEDP 0146-9592 Google Scholar
G. Schmitz and G. Muller,
“Structure and function of lamellar bodies, lipid–protein complexes involved in storage and secretion of cellular lipids,”
J. Lipid Res., 32 1539
–1570
(1991). JLPRAW 0022-2275 Google Scholar
M. Lifshitz and V. Slyozov,
“The kinetics of precipitation from supersaturated solid solutions,”
J. Phys. Chem. Solids, 19 35
–50
(1961). http://dx.doi.org/10.1016/0022-3697(61)90054-3 JPCSAW 0022-3697 Google Scholar
|